8G0M
 
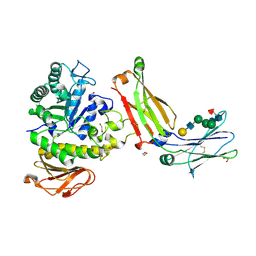 | | Structure of complex between TV6.6 and CD98hc ECD | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, TETRAETHYLENE GLYCOL, ... | | Authors: | Kariolis, M.S, Lexa, K, Liau, N.P.D, Srivastava, D, Tran, H, Wells, R.C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CD98hc is a target for brain delivery of biotherapeutics.
Nat Commun, 14, 2023
|
|
1U5Y
 
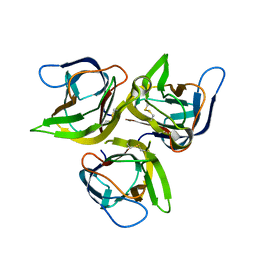 | | Crystal structure of murine APRIL, pH 8.0 | | Descriptor: | Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
1U5Z
 
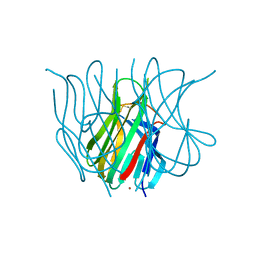 | | The Crystal structure of murine APRIL, pH 8.5 | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
1U5X
 
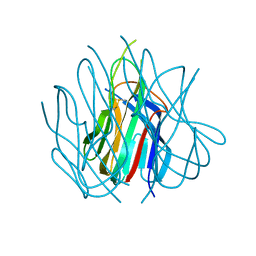 | | Crystal structure of murine APRIL at pH 5.0 | | Descriptor: | Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
5X69
 
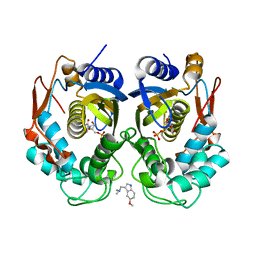 | | Human thymidylate synthase with a fragment bound in the dimer interface | | Descriptor: | 1-(5-methoxy-1H-indol-3-yl)-N,N-dimethyl-methanamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states
J. Biol. Chem., 292, 2017
|
|
4KC0
 
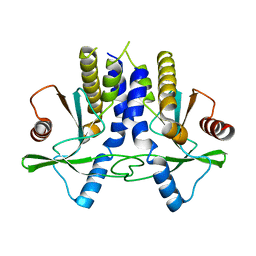 | | mSTING | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4TLH
 
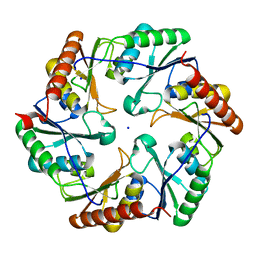 | |
4KBY
 
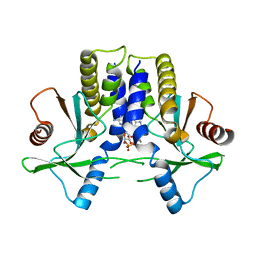 | | mSTING/c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2LE6
 
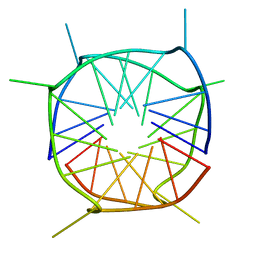 | | Structure of a dimeric all-parallel-stranded G-quadruplex stacked via the 5'-to-5' interface | | Descriptor: | DNA (5'-D(*GP*IP*GP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*T)-3') | | Authors: | Do, N.Q, Lim, K.W, Teo, M.H, Heddi, B, Phan, A.T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity
Nucleic Acids Res., 2011
|
|
7Q1Z
 
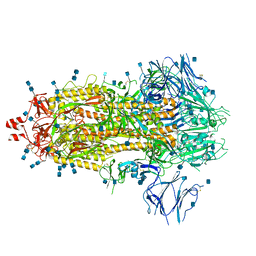 | | Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Sulbaran, G, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection.
Cell Rep Med, 3, 2022
|
|
1Z9N
 
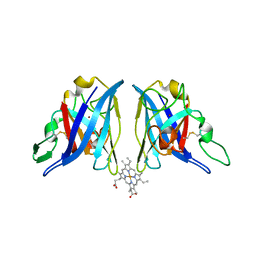 | |
1UJZ
 
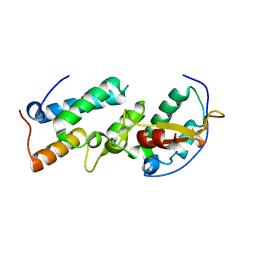 | | Crystal structure of the E7_C/Im7_C complex; a computationally designed interface between the colicin E7 DNase and the Im7 Immunity protein | | Descriptor: | Designed Colicin E7 DNase, Designed Colicin E7 immunity protein | | Authors: | Kortemme, T, Joachimiak, L.A, Bullock, A.N, Schuler, A.D, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-08-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational redesign of protein-protein interaction specificity
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1J1L
 
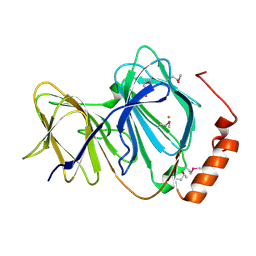 | | Crystal structure of human Pirin: a Bcl-3 and Nuclear factor I interacting protein and a cupin superfamily member | | Descriptor: | FE (II) ION, Pirin | | Authors: | Pang, H, Bartlam, M, Zeng, Q, Gao, G.F, Rao, Z. | | Deposit date: | 2002-12-10 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor
J.Biol.Chem., 279, 2004
|
|
5UPL
 
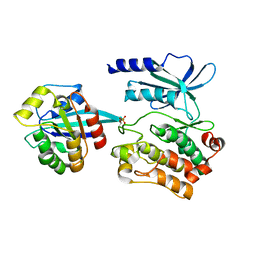 | |
5CHY
 
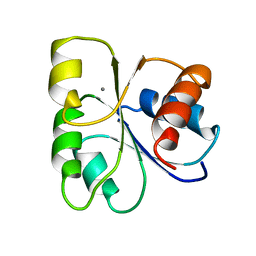 | | STRUCTURE OF CHEMOTAXIS PROTEIN CHEY | | Descriptor: | CALCIUM ION, CHEY | | Authors: | Zhu, X, Rebello, J, Matsumura, P, Volz, K. | | Deposit date: | 1996-08-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CheY mutants Y106W and T87I/Y106W. CheY activation correlates with movement of residue 106.
J.Biol.Chem., 272, 1997
|
|
3L82
 
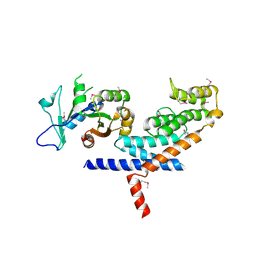 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
3SLC
 
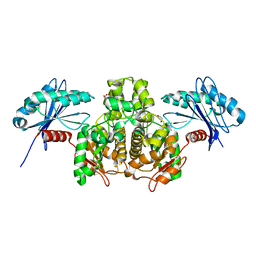 | |
1SZR
 
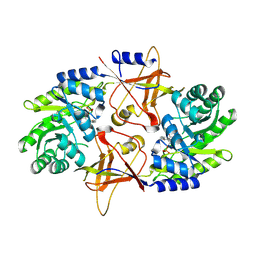 | | A Dimer interface mutant of ornithine decarboxylase reveals structure of gem diamine intermediate | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N~2~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-ORNITHINE, Ornithine decarboxylase, ... | | Authors: | Jackson, L.K, Baldwin, J, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Multiple active site conformations revealed by distant site mutation in ornithine decarboxylase
Biochemistry, 43, 2004
|
|
2R30
 
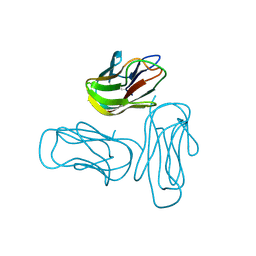 | |
3Q87
 
 | | Structure of E. cuniculi Mtq2-Trm112 complex responible for the methylation of eRF1 translation termination factor | | Descriptor: | N6 adenine specific DNA methylase, Putative uncharacterized protein ECU08_1170, S-ADENOSYLMETHIONINE, ... | | Authors: | Liger, D, Mora, L, Lazar, N, Figaro, S, Henri, J, Scrima, N, Buckingham, R.H, van Tilbeurgh, H, Heurgue-Hamard, V, Graille, M. | | Deposit date: | 2011-01-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Mechanism of activation of methyltransferases involved in translation by the Trm112 'hub' protein
Nucleic Acids Res., 2011
|
|
1EMS
 
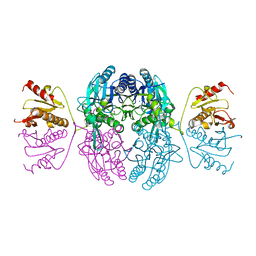 | | CRYSTAL STRUCTURE OF THE C. ELEGANS NITFHIT PROTEIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ETHYL MERCURY ION, NIT-FRAGILE HISTIDINE TRIAD FUSION PROTEIN, ... | | Authors: | Pace, H.C, Hodawadekar, S.C, Draganescu, A, Huang, J, Bieganowski, P, Pekarsky, Y, Croce, C.M, Brenner, C. | | Deposit date: | 2000-03-17 | | Release date: | 2000-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the worm NitFhit Rosetta Stone protein reveals a Nit tetramer binding two Fhit dimers.
Curr.Biol., 10, 2000
|
|
8D9V
 
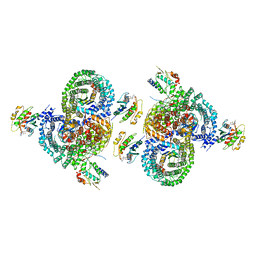 | | gamma-Arf1 homodimeric interface within AP-1, Arf1, Nef lattice on narrow membrane tubes | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8DBF
 
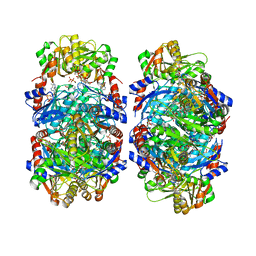 | | Human PRPS1 with ADP; Filament Interface | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBJ
 
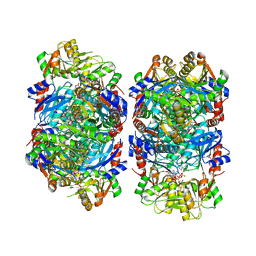 | | Human PRPS1 with Phosphate, ATP, and R5P; Filament Interface | | Descriptor: | 5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hvorecny, K.L, Kollman, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Human PRPS1 filaments stabilize allosteric sites to regulate activity.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8DBM
 
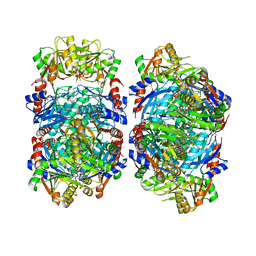 | |
