8YXK
 
 | |
8YGY
 
 | | Structure of the KLK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Kallikrein-1, SULFATE ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of the KLK1 from Biortus.
To Be Published
|
|
1HAI
 
 | | THE ISOMORPHOUS STRUCTURES OF PRETHROMBIN2, HIRUGEN-AND PPACK-THROMBIN: CHANGES ACCOMPANYING ACTIVATION AND EXOSITE BINDING TO THROMBIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Vijayalakshmi, J. | | Deposit date: | 1994-06-27 | | Release date: | 1994-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The isomorphous structures of prethrombin2, hirugen-, and PPACK-thrombin: changes accompanying activation and exosite binding to thrombin.
Protein Sci., 3, 1994
|
|
1HAO
 
 | | COMPLEX OF HUMAN ALPHA-THROMBIN WITH A 15MER OLIGONUCLEOTIDE GGTTGGTGTGGTTGG (BASED ON NMR MODEL OF DNA) | | Descriptor: | ALPHA-THROMBIN heavy chain, ALPHA-THROMBIN light chain, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Tulinsky, A, Padmanabhan, K. | | Deposit date: | 1995-10-03 | | Release date: | 1996-04-03 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An ambiguous structure of a DNA 15-mer thrombin complex.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1HX2
 
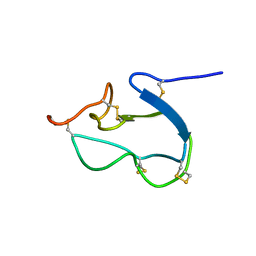 | | SOLUTION STRUCTURE OF BSTI, A TRYPSIN INHIBITOR FROM BOMBINA BOMBINA. | | Descriptor: | BSTI | | Authors: | Rosengren, K.J, Daly, N.L, Scanlon, M.J, Craik, D.J. | | Deposit date: | 2001-01-11 | | Release date: | 2001-01-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BSTI: a new trypsin inhibitor from skin secretions of Bombina bombina.
Biochemistry, 40, 2001
|
|
1TMT
 
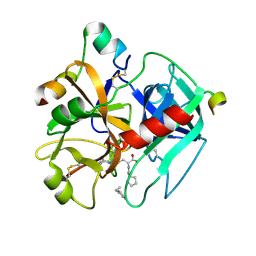 | |
7JGG
 
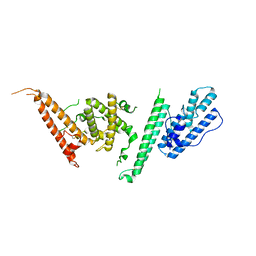 | |
7JGF
 
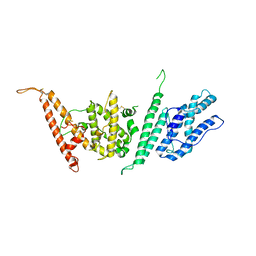 | |
7JGE
 
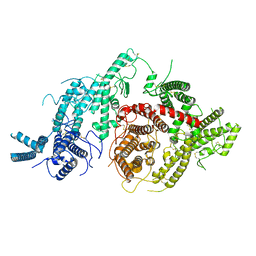 | |
7JGH
 
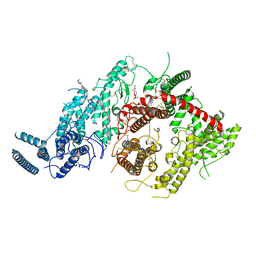 | | Cryo-EM structure of P. falciparum VAR2CSA NF54 core in complex with CSA at 3.36 A | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, Erythrocyte membrane protein 1, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid | | Authors: | Ma, R, Tolia, N.H. | | Deposit date: | 2020-07-19 | | Release date: | 2021-01-13 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for placental malaria mediated by Plasmodium falciparum VAR2CSA.
Nat Microbiol, 6, 2021
|
|
2VCP
 
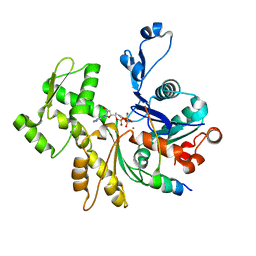 | | Crystal structure of N-Wasp VC domain in complex with skeletal actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gaucher, J.F, Didry, D, Carlier, M.F. | | Deposit date: | 2007-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interactions of isolated C-terminal fragments of neural Wiskott-Aldrich syndrome protein (N-WASP) with actin and Arp2/3 complex.
J. Biol. Chem., 287, 2012
|
|
7JGD
 
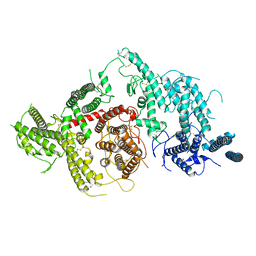 | |
7JID
 
 | | Crystal structure of the L780 UDP-rhamnose synthase from Acanthamoeba polyphaga mimivirus | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, UDP-L-Rhamnose Synthase, ... | | Authors: | Bockhaus, N.J, Ferek, J.D, Thoden, J.B, Holden, H.M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The high-resolution structure of a UDP-L-rhamnose synthase from Acanthamoeba polyphaga Mimivirus.
Protein Sci., 29, 2020
|
|
2V52
 
 | | Structure of MAL-RPEL2 complexed to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Guettler, S, Langer, C.A, Treisman, R, McDonald, N.Q. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
EMBO J., 27, 2008
|
|
7JHZ
 
 | | Crystal structure of the carbohydrate-binding domain VP8* of human P[8] rotavirus strain BM13851 in complex with LNDFH I | | Descriptor: | GLYCEROL, Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Xu, S, Stuckert, M.R, McGinnis, K.R, Jiang, X, Kennedy, M.A. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of P[II] rotavirus evolution and host ranges under selection of histo-blood group antigens.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4W6S
 
 | | Crystal Structure of Full-Length Split GFP Mutant K126C Disulfide Dimer, P 43 21 2 Space Group | | Descriptor: | GLYCEROL, PHOSPHATE ION, fluorescent protein E124H/K126C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W75
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 1 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.473 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7A
 
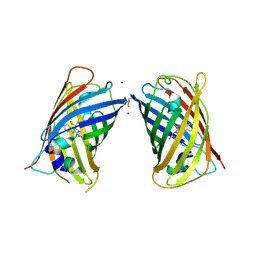 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 4 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4US6
 
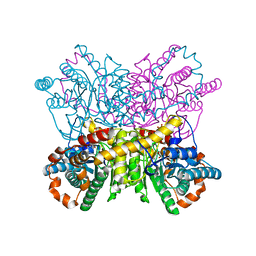 | | New Crystal Form of Glucose Isomerase Grown in Short Peptide Supramolecular Hydrogels | | Descriptor: | CALCIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Diaz-Mochon, J.J, Alvarez de Cienfuegos, L. | | Deposit date: | 2014-07-03 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Influence of the Chirality of Short Peptide Supramolecular Hydrogels in Protein Crystallogenesis.
Chem.Commun.(Camb.), 51, 2015
|
|
4V5J
 
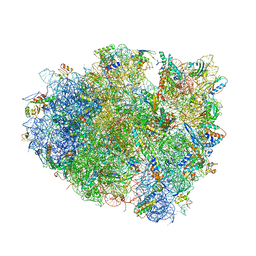 | | Structure of the 70S ribosome bound to Release factor 2 and a substrate analog provides insights into catalysis of peptide release | | Descriptor: | 16S Ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jin, H, Kelley, A.C, Loakes, D, Ramakrishnan, V. | | Deposit date: | 2010-03-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
4V5L
 
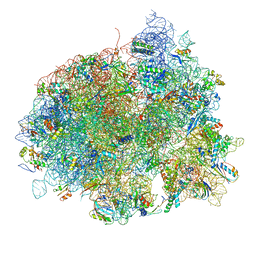 | | The structure of EF-Tu and aminoacyl-tRNA bound to the 70S ribosome with a GTP analog | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Voorhees, R.M, Schmeing, T.M, Ramakrishnan, V. | | Deposit date: | 2010-09-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Mechanism for Activation of GTP Hydrolysis on the Ribosome.
Science, 330, 2010
|
|
4UVP
 
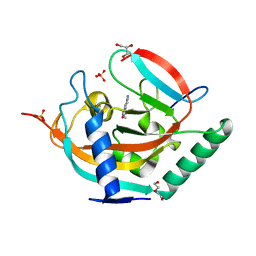 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- ethyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-ethylisoquinolin-1(2H)-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4V5N
 
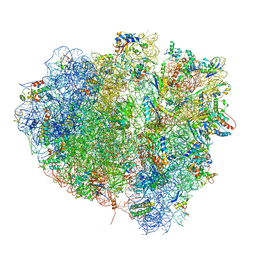 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
4V5F
 
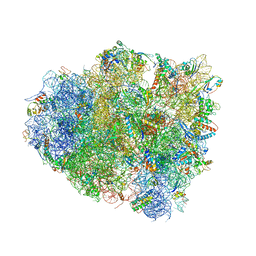 | | The structure of the ribosome with elongation factor G trapped in the post-translocational state | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.-G, Selmer, M, Dunham, C.M, Weixlbaumer, A, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-09-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of the ribosome with elongation factor G trapped in the posttranslocational state.
Science, 326, 2009
|
|
4W69
 
 | |
