2YHH
 
 | | Microvirin:mannobiose complex | | Descriptor: | MANNAN-BINDING LECTIN, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hussan, S, Bewley, C.A, Clore, G.M, Gustchina, E, Ghirlando, R. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Monovalent Lectin Microvirin in Complex with Man(Alpha)(1-2)Man Provides a Basis for Anti-HIV Activity with Low Toxicity.
J.Biol.Chem., 286, 2011
|
|
1ZGC
 
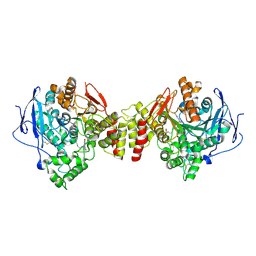 | | Crystal Structure of Torpedo Californica Acetylcholinesterase in Complex With an (RS)-Tacrine(10)-Hupyridone Inhibitor. | | Descriptor: | (5S)-5-{[10-(1,2,3,4-TETRAHYDROACRIDIN-9-YLAMINO)DECYL]AMINO}-5,6,7,8-TETRAHYDROQUINOLIN-2(1H)-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Haviv, H, Wong, D.M, Greenblatt, H.M, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-04-21 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Packing Mediates Enantioselective Ligand Recognition at the Peripheral Site of Acetylcholinesterase
J.Am.Chem.Soc., 127, 2005
|
|
3BVU
 
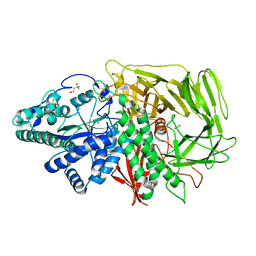 | |
4B6P
 
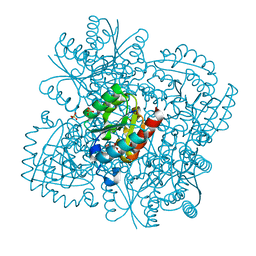 | | Structure of Mycobacterium tuberculosis Type II Dehydroquinase inhibited by (2S)-2-Perfluorobenzyl-3-dehydroquinic acid | | Descriptor: | (1R,2S,4S,5R)-2-(2,3,4,5,6-pentafluorophenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lence, E, Tizon, L, Peon, A, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic basis of the inhibition of type II dehydroquinase by (2S)- and (2R)-2-benzyl-3-dehydroquinic acids.
ACS Chem. Biol., 8, 2013
|
|
3H6U
 
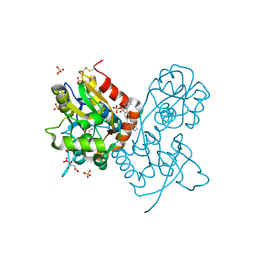 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS1493 at 1.85 A resolution | | Descriptor: | (3S)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CITRATE ANION, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
4I7N
 
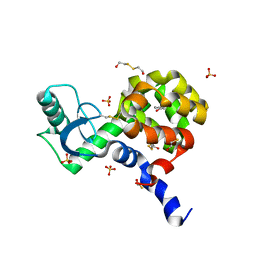 | | T4 Lysozyme L99A/M102H with 1-phenyl-2-propyn-1-ol bound | | Descriptor: | (1R)-1-phenylprop-2-yn-1-ol, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
3BVT
 
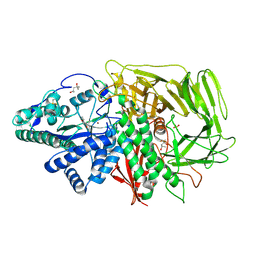 | | GOLGI MANNOSIDASE II D204A catalytic nucleophile mutant complex with Methyl (alpha-D-mannopyranosyl)-(1->3)-S-alpha-D-mannopyranoside | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-01-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the substrate specificity of Golgi alpha-mannosidase II by use of synthetic oligosaccharides and a catalytic nucleophile mutant.
J.Am.Chem.Soc., 130, 2008
|
|
4FW6
 
 | |
3NXG
 
 | | JC polyomavirus VP1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | Authors: | Neu, U, Stroeh, L.J, Stehle, T. | | Deposit date: | 2010-07-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif.
Cell Host Microbe, 8, 2010
|
|
4FAL
 
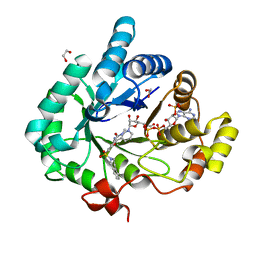 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)-N-methylbenzamide (80) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)-N-methylbenzamide, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
4RA5
 
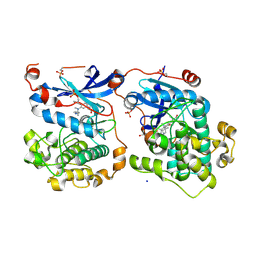 | | Human Protein Kinase C THETA IN COMPLEX WITH LIGAND COMPOUND 11a (6-[(1,3-Dimethyl-azetidin-3-yl)-methyl-amino]-4(R)-methyl-7-phenyl-2,10-dihydro-9-oxa-1,2,4a-triaza-phenanthren-3-one) | | Descriptor: | (1R)-9-[(1,3-dimethylazetidin-3-yl)(methyl)amino]-1-methyl-8-phenyl-3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, 1,2-ETHANEDIOL, HUMAN PROTEIN KINASE C THETA, ... | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimized Protein Kinase C theta (PKC theta ) Inhibitors Reveal Only Modest Anti-inflammatory Efficacy in a Rodent Model of Arthritis.
J.Med.Chem., 58, 2015
|
|
3HG2
 
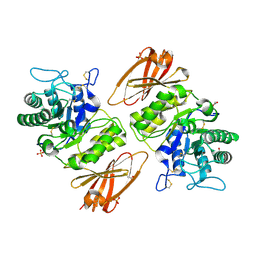 | | Human alpha-galactosidase catalytic mechanism 1. Empty active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, Alpha-galactosidase A, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
2AKZ
 
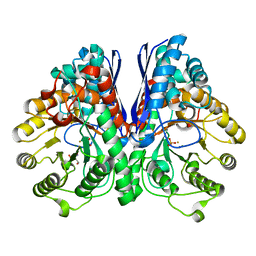 | | Fluoride Inhibition of Enolase: Crystal Structure of the Inhibitory Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLUORIDE ION, Gamma enolase, ... | | Authors: | Qin, J, Chai, G, Brewer, J.M, Lovelace, L.L. | | Deposit date: | 2005-08-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fluoride inhibition of enolase: crystal structure and thermodynamics
Biochemistry, 45, 2006
|
|
2NSL
 
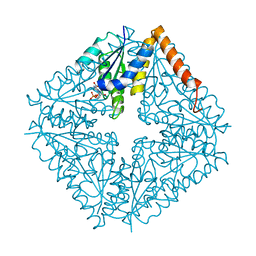 | | E. coli PurE H45N mutant complexed with CAIR | | Descriptor: | 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-11-04 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N(5)-CAIR Mutase: Role of a CO(2) Binding Site and Substrate Movement in Catalysis.
Biochemistry, 46, 2007
|
|
1TBZ
 
 | | HUMAN THROMBIN WITH ACTIVE SITE N-METHYL-D PHENYLALANYL-N-[5-(AMINOIMINOMETHYL)AMINO]-1-{{BENZOTHIAZOLYL)CARBONYL] BUTYL]-L-PROLINAMIDE TRIFLUROACETATE AND EXOSITE-HIRUGEN | | Descriptor: | ALPHA-THROMBIN, D-phenylalanyl-N-{(1S)-1-[(S)-1,3-benzothiazol-2-yl(hydroxy)methyl]-4-carbamimidamidobutyl}-L-prolinamide, HIRUGEN, ... | | Authors: | Matthews, J.H, Krishnan, R, Costanzo, M.J, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of thrombin with thiazole-containing inhibitors: probes of the S1' binding site.
Biophys.J., 71, 1996
|
|
1UWT
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with D-galactohydroximo-1,5-lactam | | Descriptor: | (2E,3R,4R,5R,6S)-3,4,5-TRIHYDROXY-6-(HYDROXYMETHYL)-2-PIPERIDINONE, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of the beta-glycosidase from Sulfolobus solfataricus in complex with covalently and noncovalently bound inhibitors.
Biochemistry, 43, 2004
|
|
2H9J
 
 | | Structure of Hen egg white lysozyme soaked with Ni2-Xylylbicyclam | | Descriptor: | 1,1'-[1,4-PHENYLENEBIS(METHYLENE)]BIS[1,4,8,11-TETRAAZACYCLOTETRADECANE]NI(II), CHLORIDE ION, Lysozyme C, ... | | Authors: | McNae, I.W, Hunter, T.M, Sadler, P.J, Walkinshaw, M.D. | | Deposit date: | 2006-06-10 | | Release date: | 2007-04-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Configurations of nickel-cyclam antiviral complexes and protein recognition.
Chemistry, 13, 2007
|
|
3QLZ
 
 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-propylpyrimidine-2,4-diamine (UCP130B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-propylpyrimidine-2,4-diamine, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
2AKM
 
 | | Fluoride Inhibition of Enolase: Crystal Structure of the Inhibitory Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gamma enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J.M, Lovelace, L.L. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fluoride inhibition of enolase: crystal structure and thermodynamics
Biochemistry, 45, 2006
|
|
7K46
 
 | |
2ALD
 
 | | HUMAN MUSCLE ALDOLASE | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE | | Authors: | Dalby, A.R, Littlechild, J.A. | | Deposit date: | 1998-10-21 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications.
Protein Sci., 8, 1999
|
|
2MPM
 
 | | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-terminal Region of CCR3 Bound to CCL11/Eotaxin-1 | | Descriptor: | CCR3, Eotaxin | | Authors: | Millard, C.J, Ludeman, J.P, Canals, M, Bridgford, J.L, Hinds, M.G, Clayton, D.J, Christopoulos, A, Payne, R.J, Stone, M.J. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: The N-Terminal Region of CCR3 Bound to CCL11/Eotaxin-1.
Structure, 22, 2014
|
|
1BBB
 
 | |
6M5B
 
 | | X-ray crystal structure of cyclic-PIP and DNA complex in a reverse binding orientation | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-[(4-azanyl-1-methyl-pyrrol-2-yl)carbonylamino]-1-methyl-pyrrol-2-yl]carbonylamino]-~{N}-[2-[[(3~{S})-3-azanyl-4-oxidanylidene-butyl]carbamoyl]-1-methyl-imidazol-4-yl]-1-methyl-imidazole-2-carboxamide, DNA (5'-D(*CP*(CBR)P*AP*GP*GP*CP*CP*TP*GP*G)-3'), ... | | Authors: | Abe, K, Hirose, Y, Eki, H, Takeda, K, Bando, T, Endo, M, Sugiyama, H. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | X-ray Crystal Structure of a Cyclic-PIP-DNA Complex in the Reverse-Binding Orientation.
J.Am.Chem.Soc., 142, 2020
|
|
5CEZ
 
 | |
