7T5P
 
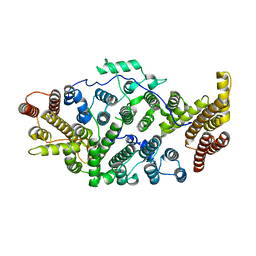 | | Cryo-EM structure of human SIMC1-SLF2 complex | | Descriptor: | SMC5-SMC6 complex localization factor protein 2, SUMO-interacting motif-containing protein 1 | | Authors: | Maeda, S, Oravcova, M, Boddy, M.N, Otomo, T. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The Nse5/6-like SIMC1-SLF2 complex localizes SMC5/6 to viral replication centers.
Elife, 11, 2022
|
|
1S1E
 
 | | Crystal Structure of Kv Channel-interacting protein 1 (KChIP-1) | | Descriptor: | CALCIUM ION, Kv channel interacting protein 1 | | Authors: | Scannevin, R.H, Wang, K.-W, Jow, F, Megules, J, Kopsco, D.C, Edris, W, Carroll, K.C, Lu, Q, Xu, W.-X, Xu, Z.-B, Katz, A.H, Olland, S, Lin, L, Taylor, M, Stahl, M, Malakian, K, Somers, W, Mosyak, L, Bowlby, M.R, Chanda, P, Rhodes, K.J. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two N-terminal domains of Kv4 K(+) channels regulate binding to and modulation by KChIP1.
Neuron, 41, 2004
|
|
1JGN
 
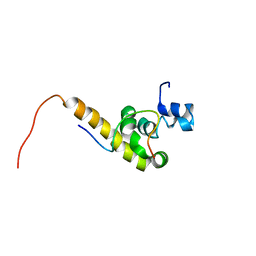 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip2 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein 2 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-26 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
5J8Z
 
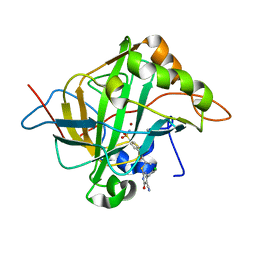 | | Human carbonic anhydrase II in complex with ligand | | Descriptor: | (2R)-2-{[4-(4-chlorophenyl)-5-cyano-6-oxo-1,6-dihydropyrimidin-2-yl]sulfanyl}-N-(4-sulfamoylphenyl)propanamide, (2S)-2-{[4-(4-chlorophenyl)-5-cyano-6-oxo-1,6-dihydropyrimidin-2-yl]sulfanyl}-N-(4-sulfamoylphenyl)propanamide, Carbonic anhydrase 2, ... | | Authors: | Alterio, V, De Simone, G. | | Deposit date: | 2016-04-08 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and X-ray crystallographic investigations of substituted 2-thio-6-oxo-1,6-dihydropyrimidine-benzenesulfonamides acting as carbonic anhydrase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
7U8R
 
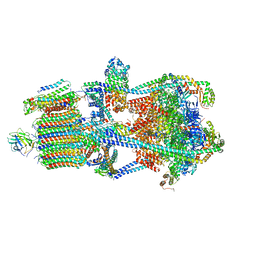 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8P
 
 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8Q
 
 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8O
 
 | | Structure of porcine V-ATPase with mEAK7 and SidK, Rotary state 2 | | Descriptor: | ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, Bacterial effector protein SidK, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
1JH4
 
 | | Solution structure of the C-terminal PABC domain of human poly(A)-binding protein in complex with the peptide from Paip1 | | Descriptor: | polyadenylate-binding protein 1, polyadenylate-binding protein-interacting protein-1 | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Ekiel, I, Gehring, K. | | Deposit date: | 2001-06-27 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
EMBO J., 23, 2004
|
|
7LR2
 
 | |
7LR7
 
 | |
7LR6
 
 | |
7LQX
 
 | |
7LR1
 
 | |
7LR8
 
 | |
6CO1
 
 | |
7KFN
 
 | | Structure of Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA containing a 2'-deoxy Benner's Base Z opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, Gli1 1W5 23mer RNA, Gli1 8AZ 23mer RNA, ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design of RNA Editing Guide Strands: Cytidine Analogs at the Orphan Position.
J.Am.Chem.Soc., 143, 2021
|
|
4ITJ
 
 | | Crystal structure of RIP1 kinase in complex with necrostatin-4 | | Descriptor: | IODIDE ION, N-[(1S)-1-(2-chloro-6-fluorophenyl)ethyl]-5-cyano-1-methyl-1H-pyrrole-2-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ITI
 
 | | Crystal structure of RIP1 kinase in complex with necrostatin-3 analog | | Descriptor: | 1-{(3S,3aS)-3-[3-fluoro-4-(trifluoromethoxy)phenyl]-8-methoxy-3,3a,4,5-tetrahydro-2H-benzo[g]indazol-2-yl}-2-hydroxyethanone, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ITH
 
 | | Crystal structure of RIP1 kinase in complex with necrostatin-1 analog | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1, ... | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
8DOV
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 (HID2) in complex with Hemoglobin | | Descriptor: | GLYCEROL, Heme-binding protein Shr, Hemoglobin subunit alpha, ... | | Authors: | Macdonald, R, Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PC6
 
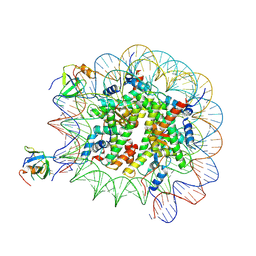 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex - pose 2 | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
3MB3
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) | | Descriptor: | 1-methylpyrrolidin-2-one, PH-interacting protein | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
8PEP
 
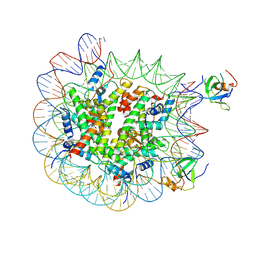 | | H3K36me2 nucleosome-LEDGF/p75 PWWP domain complex - pose 2 | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PC5
 
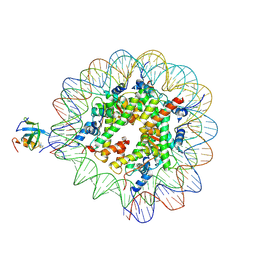 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
