3DN3
 
 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
4JTR
 
 | | AKR1C2 complex with ibuprofen | | Descriptor: | (2R)-2-[4-(2-methylpropyl)phenyl]propanoic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, ... | | Authors: | Yosaatmadja, Y, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2013-03-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of NSAID selectivity for the AKR1C family
To be Published
|
|
7E16
 
 | |
2PWG
 
 | | Crystal Structure of the Trehalulose Synthase MutB From Pseudomonas Mesoacidophila MX-45 Complexed to the Inhibitor Castanospermine | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
7E43
 
 | |
4B6G
 
 | | The Crystal Structure of the Neisserial Esterase D. | | Descriptor: | PUTATIVE ESTERASE | | Authors: | Counago, R.M, Kobe, B. | | Deposit date: | 2012-08-13 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A glutathione-dependent detoxification system is required for formaldehyde resistance and optimal survival of Neisseria meningitidis in biofilms.
Antioxid. Redox Signal., 18, 2013
|
|
7E6H
 
 | | glucose-6-phosphate dehydrogenase from Kluyveromyces lactis | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Ha, V.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
3HAZ
 
 | | Crystal structure of bifunctional proline utilization A (PutA) protein | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2009-05-03 | | Release date: | 2010-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6IQ5
 
 | | Crystal Structure of CYP1B1 and Inhibitor Having Azide Group | | Descriptor: | 2-(cis-4-azidocyclohexyl)-4H-naphtho[1,2-b]pyran-4-one, Cytochrome P450 1B1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kubo, M, Yamamoto, K, Itoh, T. | | Deposit date: | 2018-11-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design and synthesis of selective CYP1B1 inhibitor via dearomatization of alpha-naphthoflavone.
Bioorg. Med. Chem., 27, 2019
|
|
5Z18
 
 | |
3H8J
 
 | |
4JX1
 
 | | Crystal structure of reduced Cytochrome P450cam-putidaredoxin complex bound to camphor and 5-exo-hydroxycamphor | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, 5-EXO-HYDROXYCAMPHOR, CALCIUM ION, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
2GC0
 
 | |
7G2G
 
 | | Crystal Structure of rat Autotaxin in complex with 3-fluoro-4-[[4-[3-[2-[(5-methyltetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl]propanoyl]piperazin-1-yl]methyl]benzenesulfonamide, i.e. SMILES N1(CCN(CC1)Cc1c(cc(S(=O)(=O)N)cc1)F)C(=O)CCc1ccc(cc1CN1N=NC(=N1)C)C(F)(F)F with IC50=0.0203648 microM | | Descriptor: | 3-fluoro-4-{[4-(3-{2-[(5-methyl-2H-tetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl}propanoyl)piperazin-1-yl]methyl}benzene-1-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
5ZNT
 
 | | Insect chitin deacetylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | Authors: | Liu, L, Zhou, Y, Yang, Q. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
2GC7
 
 | | Substrate reduced, copper free complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans. | | Descriptor: | Amicyanin, Cytochrome c-L, HEME C, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structral comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
7G3B
 
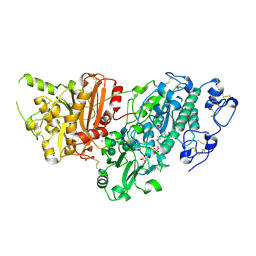 | | Crystal Structure of rat Autotaxin in complex with 3-[(2-tert-butyl-4-methylsulfonylphenoxy)methyl]-4-methyl-1H-1,2,4-triazol-5-one, i.e. SMILES O(c1ccc(cc1C(C)(C)C)S(=O)(=O)C)CC1=NNC(=O)N1C with IC50=3.84238 microM | | Descriptor: | 5-{[2-tert-butyl-4-(methanesulfonyl)phenoxy]methyl}-4-methyl-2,4-dihydro-3H-1,2,4-triazol-3-one, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G2Q
 
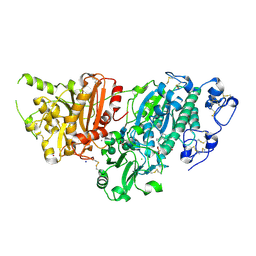 | | Crystal Structure of rat Autotaxin in complex with 1-[2-[2-cyclopropyl-6-(oxan-4-ylmethoxy)pyridine-4-carbonyl]-1,3,4,6-tetrahydropyrrolo[3,4-c]pyrrole-5-carbonyl]piperidine-4-sulfonamide, i.e. SMILES N1(C(=O)N2CC3=C(CN(C3)C(=O)c3cc(nc(c3)C3CC3)OCC3CCOCC3)C2)CC[C@@H](CC1)S(=O)(=O)N with IC50=0.00248328 microM | | Descriptor: | 1-[5-{2-cyclopropyl-6-[(oxan-4-yl)methoxy]pyridine-4-carbonyl}-3,4,5,6-tetrahydropyrrolo[3,4-c]pyrrole-2(1H)-carbonyl]piperidine-4-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
2GDW
 
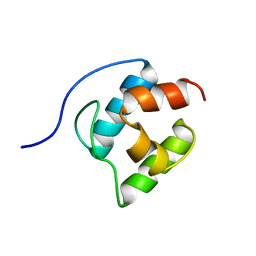 | | Solution structure of the B. brevis TycC3-PCP in A/H-state | | Descriptor: | Tyrocidine synthetase III | | Authors: | Koglin, A, Loehr, F, Rogov, V.V, Marahiel, M.A, Bernhard, F, Doetsch, V. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conformational switches modulate protein interactions in peptide antibiotic synthetases
Science, 312, 2006
|
|
7G2J
 
 | | Crystal Structure of rat Autotaxin in complex with 2-chloro-5-(5-cyclopropyloxy-2-fluorophenyl)-4-(1H-pyrazolo[3,4-b]pyridin-3-ylmethoxy)benzonitrile, i.e. SMILES C1C(C1)Oc1ccc(F)c(c1)c1cc(C#N)c(Cl)cc1OCC1=NNc2ncccc12 with IC50=0.0201566 microM | | Descriptor: | (1P)-4-chloro-5'-(cyclopropyloxy)-2'-fluoro-6-[(1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy][1,1'-biphenyl]-3-carbonitrile, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
4OHF
 
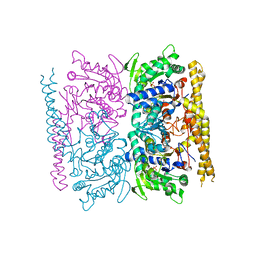 | | Crystal structure of cytosolic nucleotidase II (LPG0095) in complex with GMP from Legionella pneumophila, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET LGR1 | | Descriptor: | Cytosolic IMP-GMP specific 5'-nucleotidase, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Srinivisan, B, Forouhar, F, Shukla, A, Sampangi, C, Kulkarni, S, Abashidze, M, Seetharaman, J, Lew, S, Mao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.M, Tong, L, Balaram, H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila.
Febs J., 281, 2014
|
|
7G3M
 
 | | Crystal Structure of rat Autotaxin in complex with 2-[(2-tert-butyl-5-chloro-4-cyanophenoxy)methyl]-3-methyl-N-(oxetan-3-yl)imidazole-4-carboxamide, i.e. SMILES Clc1c(C#N)cc(C(C)(C)C)c(c1)OCC1=NC=C(C(=O)NC2COC2)N1C with IC50=0.0161695 microM | | Descriptor: | 2-[(2-tert-butyl-5-chloro-4-cyanophenoxy)methyl]-1-methyl-N-(oxetan-3-yl)-1H-imidazole-5-carboxamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Green, L, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G3R
 
 | | Crystal Structure of rat Autotaxin in complex with 5-tert-butyl-2-methyl-4-[(6-oxo-4,5-dihydro-1H-pyridazin-3-yl)methoxy]benzonitrile, i.e. SMILES O=C1NN=C(CC1)COc1cc(C)c(C#N)cc1C(C)(C)C with IC50=0.0299365 microM | | Descriptor: | 5-tert-butyl-2-methyl-4-[(6-oxo-1,4,5,6-tetrahydropyridazin-3-yl)methoxy]benzonitrile, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Ratni, H, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G2N
 
 | | Crystal Structure of rat Autotaxin in complex with 5-fluoro-6-[1-[3-[2-[(5-methyltetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl]propanoyl]piperidin-4-yl]sulfanylpyridine-3-sulfonamide, i.e. SMILES O=C(N1CC[C@@H](Sc2c(F)cc(S(=O)(=O)N)cn2)CC1)CCc1ccc(cc1CN1N=NC(=N1)C)C(F)(F)F with IC50=0.00881095 microM | | Descriptor: | 5-fluoro-6-{[1-(3-{2-[(5-methyl-2H-tetrazol-2-yl)methyl]-4-(trifluoromethyl)phenyl}propanoyl)piperidin-4-yl]sulfanyl}pyridine-3-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3HCJ
 
 | |
