5W1I
 
 | |
3EQT
 
 | |
5TDK
 
 | | RNA decamer duplex with eight 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
4OOW
 
 | | HCV NS5B polymerase with a fragment of quercetagetin | | Descriptor: | CATECHOL, RNA-directed RNA polymerase | | Authors: | Guichou, J.F, Ahmed-Belkacem, A, Rozenn, B, Nazim, N, Hernandez, E, Pallier, C, Pawlotsky, J.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: a new mechanism for antiviral intervention.
Nucleic Acids Res., 42, 2014
|
|
5VJ9
 
 | |
5TDJ
 
 | | RNA decamer duplex with four 2'-5'-linkages | | Descriptor: | RNA (5'-R(*CP*CP*GP*GP*CP*GP*CP*CP*GP*G)-3'), STRONTIUM ION | | Authors: | Luo, Z, Sheng, J. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into RNA duplexes with multiple 2 -5 -linkages.
Nucleic Acids Res., 45, 2017
|
|
8ITS
 
 | |
3OTD
 
 | | Crystal structure of human tRNAHis guanylyltransferase (Thg1)- NaI derivative | | Descriptor: | IODIDE ION, tRNA(His) guanylyltransferase | | Authors: | Hyde, S.J, Eckenroth, B.E, Doublie, S. | | Deposit date: | 2010-09-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8T1R
 
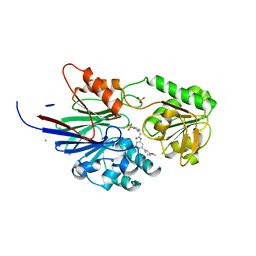 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 2 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(3-methoxyphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
8T1Q
 
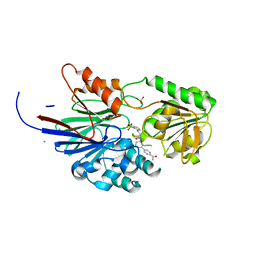 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
5UZ9
 
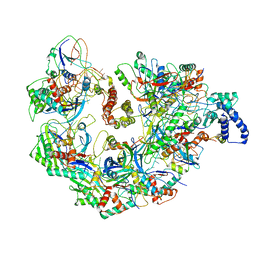 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
1Y39
 
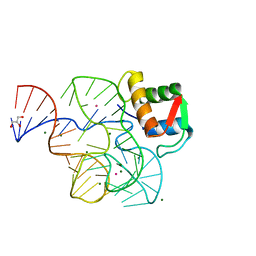 | | Co-evolution of protein and RNA structures within a highly conserved ribosomal domain | | Descriptor: | 50S ribosomal protein L11, 58 Nucleotide Ribosomal 23S RNA Domain, COBALT (III) ION, ... | | Authors: | Dunstan, M.S, GuhaThakurta, D, Draper, D.E, Conn, G.L. | | Deposit date: | 2004-11-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coevolution of Protein and RNA Structures within a Highly Conserved Ribosomal Domain
Chem.Biol., 12, 2005
|
|
2NR0
 
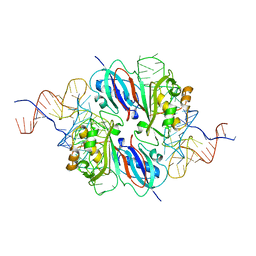 | |
6BU9
 
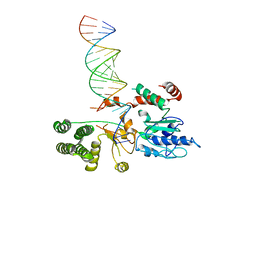 | | Drosophila Dicer-2 bound to blunt dsRNA | | Descriptor: | Dicer-2, isoform A, RNA (5'-R(*AP*CP*UP*AP*CP*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*C)-3'), ... | | Authors: | Shen, P.S, Sinha, N.K, Bass, B.L. | | Deposit date: | 2017-12-09 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Dicer uses distinct modules for recognizing dsRNA termini.
Science, 359, 2018
|
|
8R3Z
 
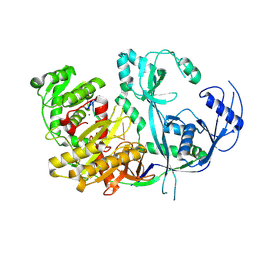 | |
3WBM
 
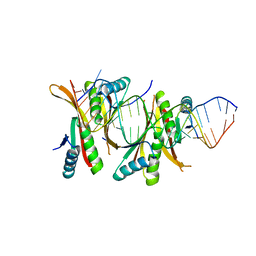 | | Crystal Structure of protein-RNA complex | | Descriptor: | DNA/RNA-binding protein Alba 1, RNA (25-MER) | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea.
J.Biol.Chem., 289, 2014
|
|
6XMG
 
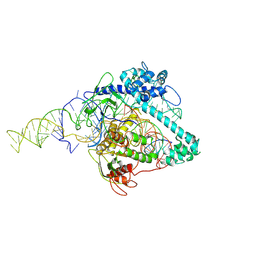 | | Cryo-EM structure of Cas12g ternary complex | | Descriptor: | CRISPR-Cas, RNA (130-MER), RNA (5'-R(P*UP*UP*AP*AP*UP*GP*CP*GP*GP*UP*AP*GP*UP*UP*UP*AP*UP*CP*AP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2020-06-30 | | Release date: | 2021-01-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of the RNA-guided ribonuclease Cas12g.
Nat.Chem.Biol., 17, 2021
|
|
3LTI
 
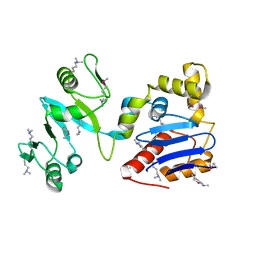 | |
4NRU
 
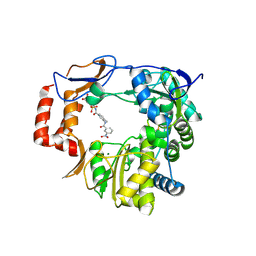 | | Murine Norovirus RNA-dependent-RNA-polymerase in complex with Compound 6, a suramin derivative | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, MAGNESIUM ION, RNA dependent RNA polymerase | | Authors: | Milani, M, Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
7QP2
 
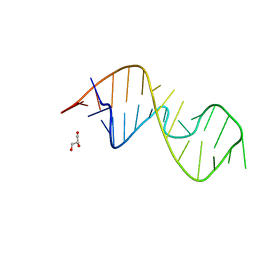 | | 1-deazaguanosine modified-RNA Sarcin Ricin Loop | | Descriptor: | GLYCEROL, RNA (27-MER) | | Authors: | Ennifar, E, Micura, R, Bereiter, R, Renard, E, Kreutz, C. | | Deposit date: | 2021-12-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | 1-Deazaguanosine-Modified RNA: The Missing Piece for Functional RNA Atomic Mutagenesis.
J.Am.Chem.Soc., 144, 2022
|
|
5CD4
 
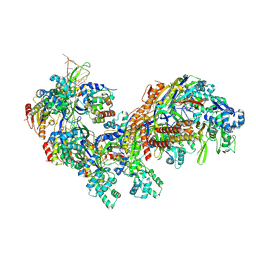 | | The Type IE CRISPR Cascade complex from E. coli, with two assemblies in the asymmetric unit arranged back-to-back | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2015-07-03 | | Release date: | 2015-08-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
6EOJ
 
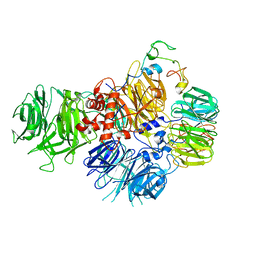 | | PolyA polymerase module of the cleavage and polyadenylation factor (CPF) from Saccharomyces cerevisiae | | Descriptor: | Polyadenylation factor subunit 2,Polyadenylation factor subunit 2, Protein CFT1, ZINC ION, ... | | Authors: | Casanal, A, Kumar, A, Hill, C.H, Emsley, P, Passmore, L.A. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Architecture of eukaryotic mRNA 3'-end processing machinery.
Science, 358, 2017
|
|
7Z26
 
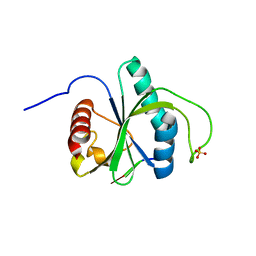 | | Crystal structure of YTHDF2 YTH domain in complex with m6A RNA | | Descriptor: | GLYCEROL, RNA (5'-R(P*(6MZ)P*CP*U)-3'), SULFATE ION, ... | | Authors: | Nai, F, Nachawati, R, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
4MJ7
 
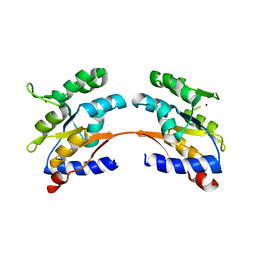 | |
6HBX
 
 | |
