8EI0
 
 | | Crystal structure of the STUB1 TPR domain in complex with H318, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, H318, ... | | Authors: | Li, K, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI3
 
 | | Crystal structure of VHL in complex with H313, a Helicon Polypeptide | | Descriptor: | Elongin-B, Elongin-C, H313, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
6N9P
 
 | |
8EI6
 
 | | Crystal structure of the WWP2 HECT domain in complex with H305, a Helicon Polypeptide | | Descriptor: | H305, N,N'-(1,4-phenylene)diacetamide, NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EI5
 
 | | Crystal structure of the WWP2 HECT domain in complex with H301, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, H301, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
6MWM
 
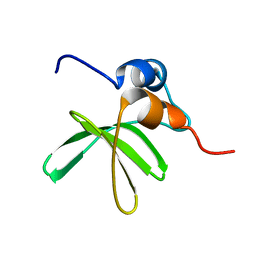 | | Bat coronavirus HKU4 SUD-C | | Descriptor: | Non-structural protein 3 | | Authors: | Staup, A.J, De Silva, I.U, Catt, J.T, Tan, X, Hammond, R.G, Johnson, M.A. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the SARS-Unique Domain C From the Bat Coronavirus HKU4.
Nat Prod Commun, 14, 2019
|
|
6MK5
 
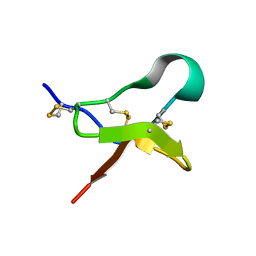 | |
8EI4
 
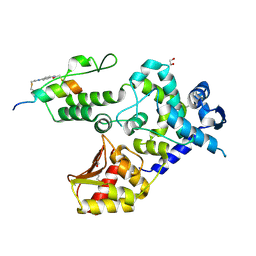 | | Crystal structure of the WWP1 HECT domain in complex with H302, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, H302, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8EIA
 
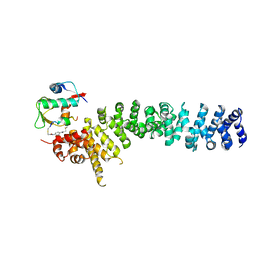 | | Crystal structure of beta-catenin and the MDM2 p53-binding domain in complex with H333, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, E3 ubiquitin-protein ligase Mdm2, H333, ... | | Authors: | Li, K, Travaline, T.L, Swiecicki, J.-M, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
8F1F
 
 | | Structure of K48-linked tri-ubiquitin in complex with cyclic peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Non-proteinogenic cyclic peptide (inhibitor), ... | | Authors: | Lubkowski, J, Fushman, D, Lemma, B. | | Deposit date: | 2022-11-05 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
6G2R
 
 | | Crystal structure of FimH in complex with a tetraflourinated biphenyl alpha D-mannoside | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]-2,3,5,6-tetrakis(fluoranyl)benzenecarbonitrile, SULFATE ION, ... | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
8F7Z
 
 | | VRC34.01_mm28 bound to fusion peptide | | Descriptor: | HIV-1 Env Fusion Peptide, VRC34_m228 Light Chain, VRC34_mm28 Heavy Chain | | Authors: | Olia, A.S, Kwong, P.D. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
6NEM
 
 | |
3T4N
 
 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
4ODP
 
 | | Structure of SlyD delta-IF from Thermus thermophilus in complex with S2-W23A peptide | | Descriptor: | 30S ribosomal protein S2, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
8ESS
 
 | | Myoglobin variant Mb-cIII complex | | Descriptor: | Myoglobin, SULFATE ION, [2,18-bis(2-carboxyethyl)-7,12-diethenyl-3,8,13,17-tetramethyl-21-(2-oxo-3-phenylpropyl)porphyrin-21-iumato(2-)-kappa~3~N~22~,N~23~,N~24~]iron(2+) | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
8ESU
 
 | | Myoglobin variant Mb-imi complex | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
1NA8
 
 | | Crystal structure of ADP-ribosylation factor binding protein GGA1 | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Lui, W.W, Collins, B.M, Hirst, J, Motley, A, Millar, C, Schu, P, Owen, D.J, Robinson, M.S. | | Deposit date: | 2002-11-27 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding partners for the COOH-terminal appendage domains of the GGAs and gamma-adaptin
Mol.Cell.Biol., 14, 2003
|
|
8ENX
 
 | |
8ENS
 
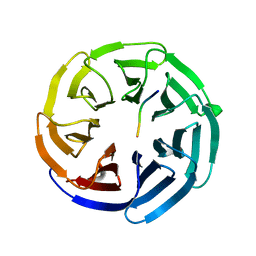 | |
8ENW
 
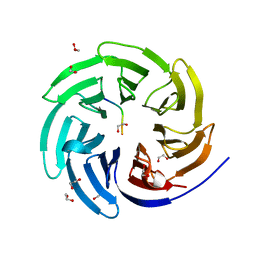 | |
4ODO
 
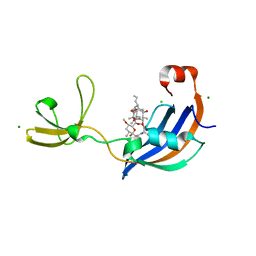 | | Structure of SlyD from Thermus thermophilus in complex with FK506 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
6NEL
 
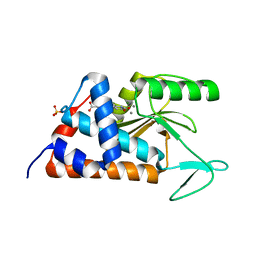 | | 4-(2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzoic acid bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 4-[2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl]benzoic acid, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aryl and Arylalkyl Substituted 3-Hydroxypyridin-2(1H)-ones: Synthesis and Evaluation as Inhibitors of Influenza A Endonuclease.
Chemmedchem, 14, 2019
|
|
8FMX
 
 | | Langya virus F glycoprotein ectodomain in prefusion form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Low, Y.S, Isaacs, A, Modhiran, N, Watterson, D. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure and antigenicity of divergent Henipavirus fusion glycoproteins.
Nat Commun, 14, 2023
|
|
8FMY
 
 | |
