1MAI
 
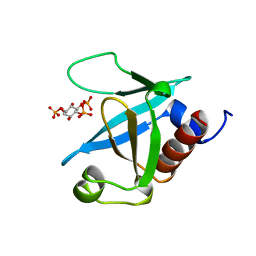 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM PHOSPHOLIPASE C DELTA IN COMPLEX WITH INOSITOL TRISPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOLIPASE C DELTA-1 | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1996-05-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain.
Cell(Cambridge,Mass.), 83, 1995
|
|
4HOK
 
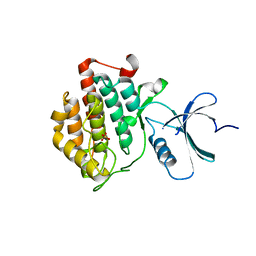 | | crystal structure of apo ck1e | | Descriptor: | Casein kinase I isoform epsilon, SULFATE ION | | Authors: | Huang, X, Long, A.M, Zhao, H. | | Deposit date: | 2012-10-22 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for the potent and selective inhibition of casein kinase 1 epsilon.
J.Med.Chem., 55, 2012
|
|
4HR7
 
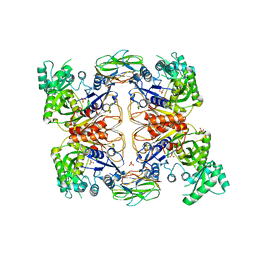 | | Crystal Structure of Biotin Carboxyl Carrier Protein-Biotin Carboxylase Complex from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxyl carrier protein of acetyl-CoA carboxylase, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Kobe, M.J, Pakhomova, S, Neau, D.B, Price, A.E, Champion, T.S, Waldrop, G.L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase.
Structure, 21, 2013
|
|
2PM1
 
 | |
1QL0
 
 | |
1VSO
 
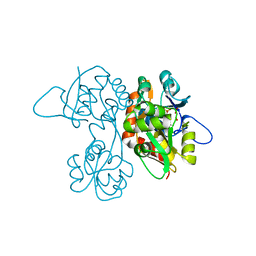 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | Authors: | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-03-29 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
1BAM
 
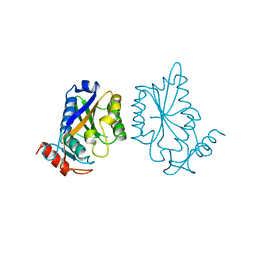 | |
4KX5
 
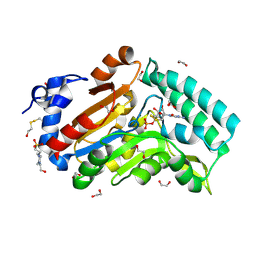 | |
5PP1
 
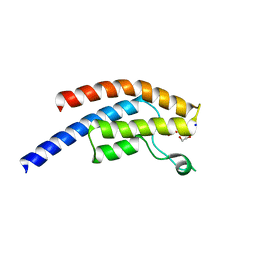 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 1) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1R5S
 
 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | Authors: | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.357 Å) | | Cite: | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
3CQU
 
 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[2-(5-methyl-4H-1,2,4-triazol-3-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1WQ9
 
 | | Crystal structure of VR-1, a VEGF-F from a snake venom | | Descriptor: | Vascular endothelial growth factor | | Authors: | Suto, K, Yamazaki, Y, Morita, T, Mizuno, H. | | Deposit date: | 2004-09-24 | | Release date: | 2004-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of novel vascular endothelial growth factors (VEGF) from snake venoms: insight into selective VEGF binding to kinase insert domain-containing receptor but not to fms-like tyrosine kinase-1.
J.Biol.Chem., 280, 2005
|
|
1ZRH
 
 | | Crystal structure of Human heparan sulfate glucosamine 3-O-sulfotransferase 1 in complex with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 1 | | Authors: | Dong, A, Dombrovski, L, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Human heparan sulfate glucosamine 3-O-sulfotransferase 1 in complex with PAP
To be Published
|
|
4LI7
 
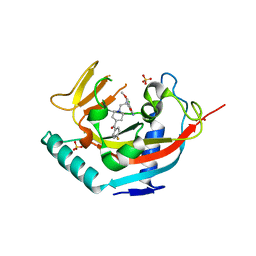 | | TANKYRASE-1 complexed with small molecule inhibitor 4-chloro-5-cyano-N-{2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl}-2-methoxybenzamide | | Descriptor: | 4-chloro-5-cyano-N-{2-[4-(4-fluorobenzoyl)piperidin-1-yl]ethyl}-2-methoxybenzamide, SODIUM ION, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
5YTS
 
 | | Crystal structure of YB1 cold-shock domain in complex with UCUUCU | | Descriptor: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*CP*UP*UP*C)-3'), SULFATE ION | | Authors: | Yang, X, Huang, Y. | | Deposit date: | 2017-11-20 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA.
J.Biol.Chem., 294, 2019
|
|
1Q1J
 
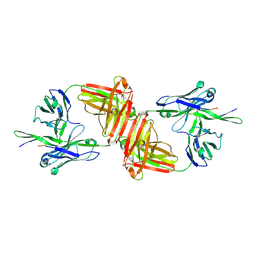 | | Crystal Structure Analysis of anti-HIV-1 Fab 447-52D in complex with V3 peptide | | Descriptor: | Fab 447-52D, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Williams, C, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2003-07-21 | | Release date: | 2004-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D.
Structure, 12, 2004
|
|
4AV0
 
 | | Structure of the FimH lectin domain in the trigonal space group, in complex with a methoxy phenyl propynyl alpha-D-mannoside at 2.1 A resolution | | Descriptor: | 3-(4-methoxyphenyl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
1X45
 
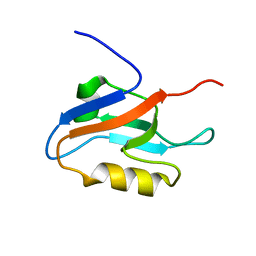 | | Solution structure of the first PDZ domain of amyloid beta A4 precursor protein-binding family A, member 1 | | Descriptor: | amyloid beta (A4) precursor protein-binding, family A, member 1 (X11) | | Authors: | Qin, X.R, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-13 | | Release date: | 2005-11-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first PDZ domain of amyloid beta A4 precursor protein-binding family A, member 1
to be published
|
|
1PKR
 
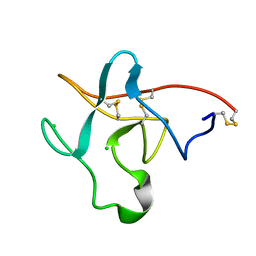 | |
3MBK
 
 | |
1ZUR
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1F) | | Descriptor: | CHLORIDE ION, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-4-phenyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-05-31 | | Release date: | 2006-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R1F)
To be Published
|
|
1PPL
 
 | |
6LZZ
 
 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 4a | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(2-oxa-6-azaspiro[3.3]heptan-6-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, Y.Y, Wu, Y, Luo, H.B. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.40003753 Å) | | Cite: | Identification of phosphodiesterase-9 as a novel target for pulmonary arterial hypertension by using highly selective and orally bioavailable inhibitors
To Be Published
|
|
3WF8
 
 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate | | Descriptor: | 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
