7TBL
 
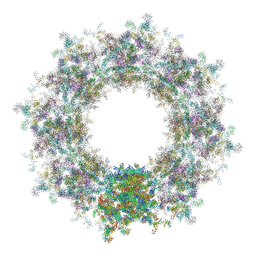 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | DDX19, ELYS, GLE1, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7AY0
 
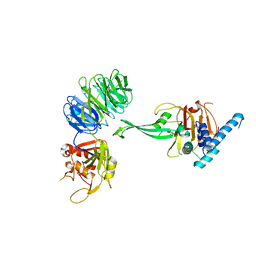 | | Crystal structure of truncated USP1-UAF1 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 1, WD repeat-containing protein 48, ZINC ION | | Authors: | Arkinson, C, Rennie, M.L, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2IHB
 
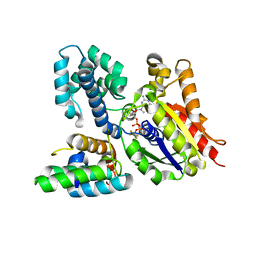 | | Crystal structure of the heterodimeric complex of human RGS10 and activated Gi alpha 3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, MAGNESIUM ION, ... | | Authors: | Soundararajan, M, Turnbull, A.P, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Ugochukwu, E, Gorrec, F, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7KHL
 
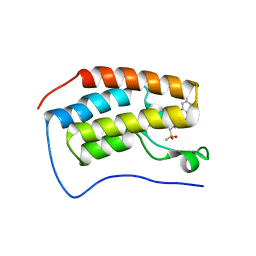 | | BRD4-BD1 Compound6 (methyl 4-(3,5-difluoropyridin-2-yl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxylate) | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, methyl 7-(3,5-difluoropyridin-2-yl)-2-methyl-10-[(methylsulfonyl)methyl]-3-oxo-3,4,6,7-tetrahydro-2H-2,4,7-triazadibenzo[cd,f]azulene-9-carboxylate | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.286 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
8Q9L
 
 | |
7L5Q
 
 | | The empty AAV7 capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2020-12-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Completion of the AAV Structural Atlas: Serotype Capsid Structures Reveals Clade-Specific Features.
Viruses, 13, 2021
|
|
8PE8
 
 | |
7Q4W
 
 | | CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Poeverlein, M.C, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
2M4E
 
 | | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333 | | Descriptor: | Putative uncharacterized protein | | Authors: | Wu, B, Yee, A, Houliston, S, Lemak, A, Garcia, M, Savchenko, A, Arrowsmith, C.H, Anderson, W.F, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VV2_0175 from Vibrio vulnificus, NESG target VnR1 and CSGID target IDP91333
To be Published
|
|
3IQG
 
 | |
2M3C
 
 | | Solution Structure of gammaM7-Crystallin | | Descriptor: | Crystallin, gamma M7 | | Authors: | Mahler, B, Wu, Z. | | Deposit date: | 2013-01-16 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Fish Eye Lens Protein, gamma M7-Crystallin.
Biochemistry, 52, 2013
|
|
7L6N
 
 | | The Mycobacterium tuberculosis ClpB disaggregase hexamer structure with three locally refined ClpB middle domains and three DnaK nucleotide binding domains | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB, Chaperone protein DnaK, ... | | Authors: | Yin, Y.Y, Feng, X, Li, H. | | Deposit date: | 2020-12-23 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for aggregate dissolution and refolding by the Mycobacterium tuberculosis ClpB-DnaK bi-chaperone system.
Cell Rep, 35, 2021
|
|
7Q4V
 
 | | Electron bifurcating hydrogenase - HydABC from A. woodii | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Katsyv, A, Kumar, A, Saura, P, Poeverlein, M.C, Freibert, S.A, Stripp, S, Jain, S, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2021-11-02 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
5KZ7
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 7-[(1~{S})-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(pyridin-3-ylamino)pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7AY2
 
 | | Crystal structure of truncated USP1-UAF1 reacted with ubiquitin-prg | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 1, WD repeat-containing protein 48, ... | | Authors: | Arkinson, C, Rennie, M.L, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0Z
 
 | | Spinach variant bound to DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Truong, L, Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2020-12-13 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fluorogenic aptamers resolve the flexibility of RNA junctions using orientation-dependent FRET.
Rna, 27, 2021
|
|
5KZF
 
 | |
7PU4
 
 | |
5L1O
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
7PSC
 
 | | Crystal structure of the disease-causing I358T mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Nemes-Nikodem, E, Szabo, E, Zambo, Z, Vass, K.R, Taberman, H, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2021-09-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Structural and Biochemical Investigation of Selected Pathogenic Mutants of the Human Dihydrolipoamide Dehydrogenase.
Int J Mol Sci, 24, 2023
|
|
7SSX
 
 | | Structure of human Kv1.3 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
3IRK
 
 | | Solution Structure of Heparin dp30 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose | | Authors: | Khan, S, Gor, J, Mulloy, B, Perkins, S.J. | | Deposit date: | 2009-08-24 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin-protein complexes.
J.Mol.Biol., 395, 2010
|
|
7SSY
 
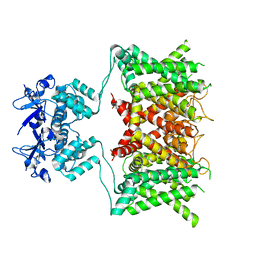 | | Structure of human Kv1.3 (alternate conformation) | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
8Q7L
 
 | |
7SSZ
 
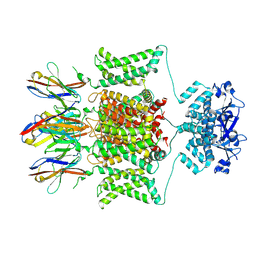 | | Structure of human Kv1.3 with A0194009G09 nanobodies | | Descriptor: | Nanobody A0194009G09, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3,Green fluorescent protein fusion | | Authors: | Meyerson, J.R, Selvakumar, P. | | Deposit date: | 2021-11-11 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
