2FB6
 
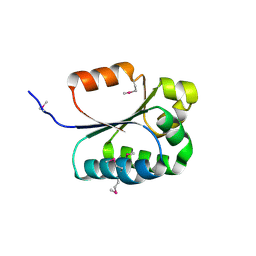 | | Structure of Conserved Protein of Unknown Function BT1422 from Bacteroides thetaiotaomicron | | Descriptor: | conserved hypothetical protein | | Authors: | Osipiuk, J, Mulligan, R, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-08 | | Release date: | 2006-01-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of conserved hypothetical protein BT1422 from Bacteroides thetaiotaomicron
To be Published
|
|
2FEE
 
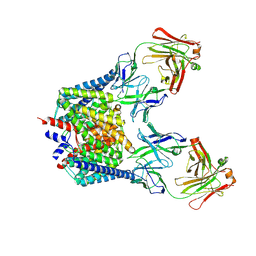 | | Structure of the Cl-/H+ exchanger CLC-ec1 from E.Coli in NaBr | | Descriptor: | Fab fragment, heavy chain, light chain, ... | | Authors: | Accardi, A, Walden, M.P, Nguitragool, W, Jayaram, H, Williams, C, Miller, C. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Separate ion pathways in a Cl-/H+ exchanger
J.Gen.Physiol., 126, 2005
|
|
6W3W
 
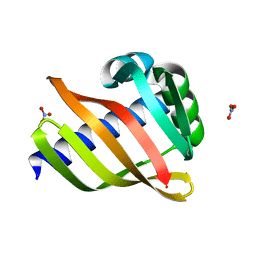 | | An enumerative algorithm for de novo design of proteins with diverse pocket structures | | Descriptor: | DENOVO NTF2, NITRATE ION | | Authors: | Bera, A.K, Basanta, B, Dimaio, F, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5H88
 
 | | Crystal structure of mRojoA mutant - T16V -P63F - W143A - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5HAR
 
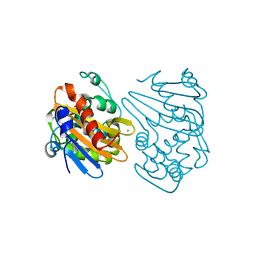 | | OXA-163 beta-lactamase - S70G mutant | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION | | Authors: | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
5H89
 
 | | Crystal structure of mRojoA mutant - T16V - P63Y - W143G - L163V | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
2F4J
 
 | | Structure of the Kinase Domain of an Imatinib-Resistant Abl Mutant in Complex with the Aurora Kinase Inhibitor VX-680 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Young, M.A, Shah, N.P, Chao, L.H, Zarrinkar, P, Sawyers, P, Kuriyan, J. | | Deposit date: | 2005-11-23 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680.
Cancer Res., 66, 2006
|
|
6MXR
 
 | |
2FTL
 
 | | Crystal structure of trypsin complexed with BPTI at 100K | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
2FZU
 
 | | Reduced enolate chromophore intermediate for GFP variant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural evidence for an enolate intermediate in GFP fluorophore biosynthesis.
J.Am.Chem.Soc., 128, 2006
|
|
2FUV
 
 | | Phosphoglucomutase from Salmonella typhimurium. | | Descriptor: | MAGNESIUM ION, phosphoglucomutase | | Authors: | Osipiuk, J, Wu, R, Holzle, D, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-27 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of phosphoglucomutase from Salmonella typhimurium.
To be Published
|
|
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | Descriptor: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | Authors: | Shi, R, Picard, M.-E, Manenda, M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
2G5P
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 21ac | | Descriptor: | 4-{[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHOXY}-3-TERT-BUTYLBENZOIC ACID, Dipeptidyl peptidase 4 | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-23 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
2G63
 
 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) complexed with cyanopyrrolidine (C5-pro-pro) inhibitor 24b | | Descriptor: | Dipeptidyl peptidase 4, METHYL 4-{[({[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHYL}AMINO)CARBONYL]AMINO}BENZOATE | | Authors: | Longenecker, K.L, Fry, E.H, Lake, M.R, Solomon, L.R, Pei, Z, Li, X. | | Deposit date: | 2006-02-24 | | Release date: | 2006-07-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery, structure-activity relationship, and pharmacological evaluation of (5-substituted-pyrrolidinyl-2-carbonyl)-2-cyanopyrrolidines as potent dipeptidyl peptidase IV inhibitors.
J.Med.Chem., 49, 2006
|
|
5HAI
 
 | | P99 beta-lactamase mutant - S64G | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Stojanoski, V, Adamski, C.J, Hu, L, Mehta, S.C, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Removal of the Side Chain at the Active-Site Serine by a Glycine Substitution Increases the Stability of a Wide Range of Serine beta-Lactamases by Relieving Steric Strain.
Biochemistry, 55, 2016
|
|
6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3G
 
 | | Rd1NTF2_04 with long sheet | | Descriptor: | Rd1NTF2_04 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5H87
 
 | | Crystal structure of mRojoA mutant - P63H - W143S | | Descriptor: | mRojoA fluorescent protein | | Authors: | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
5GLC
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR11 containing 20 residues insertion in omega-loop | | Descriptor: | Beta-lactamase | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLD
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR11 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GIT
 
 | | BTB domain of KEAP1 in complex with XX3 | | Descriptor: | Kelch-like ECH-associated protein 1, [(3aS,5R,5aS,6S,8S,8aS,9S,9aR)-9-acetyloxy-8-hydroxy-5,8a-dimethyl-1-methylidene-2-oxo-4,5,5a,6,7,8,9,9a-octahydro-3aH-azuleno[6,5-b]furan-6-yl] acetate | | Authors: | Zhu, L.L, Li, H.L, Wu, F.S, Xiong, R. | | Deposit date: | 2016-06-25 | | Release date: | 2017-06-28 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Britanin Ameliorates Cerebral Ischemia-Reperfusion Injury by Inducing the Nrf2 Protective Pathway.
Antioxid. Redox Signal., 27, 2017
|
|
5GOT
 
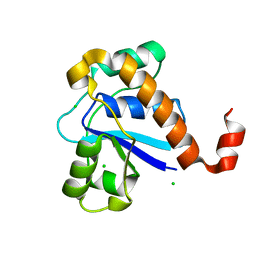 | | Crystal structure of SP-PTP, low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, Low molecular weight phosphotyrosine phosphatase family protein | | Authors: | Ku, B, Keum, C.W, KIim, S.J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of SP-PTP, a low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes
Biochem.Biophys.Res.Commun., 478, 2016
|
|
2FX3
 
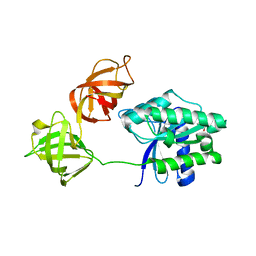 | | Crystal Structure Determination of E. coli Elongation Factor, Tu using a Twinned Data Set | | Descriptor: | Elongation factor Tu, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Heffron, S.E, Moeller, R, Jurnak, F. | | Deposit date: | 2006-02-03 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Solving the structure of Escherichia coli elongation factor Tu using a twinned data set.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2G4C
 
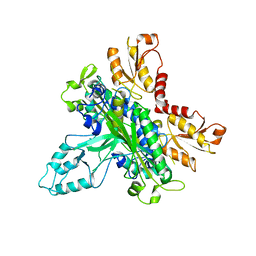 | | Crystal Structure of human DNA polymerase gamma accessory subunit | | Descriptor: | DNA polymerase gamma subunit 2 | | Authors: | Fan, L, Farr, C.L, Kaguni, L.S, Tainer, J.A. | | Deposit date: | 2006-02-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A novel processive mechanism for DNA synthesis revealed by structure, modeling and mutagenesis of the accessory subunit of human mitochondrial DNA polymerase
J.Mol.Biol., 358, 2006
|
|
5HFN
 
 | |
