2JCH
 
 | | Structural and mechanistic basis of penicillin binding protein inhibition by lactivicins | | Descriptor: | (2E)-2-({(2S)-2-CARBOXY-2-[(PHENOXYACETYL)AMINO]ETHOXY}IMINO)PENTANEDIOIC ACID, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2006-12-23 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
4FHA
 
 | | Structure of Dihydrodipicolinate Synthase from Streptococcus pneumoniae,bound to pyruvate and lysine | | Descriptor: | Dihydrodipicolinate synthase, LYSINE, SODIUM ION | | Authors: | Perugini, M.A, Dogovski, C, Parker, M.W, Gorman, M.A. | | Deposit date: | 2012-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure, Function, Stability and Knockout Phenotype of Dihydrodipicolinate Synthase from Streptococcus pneumoniae
To be Published
|
|
6MCC
 
 | |
4FF5
 
 | |
4HFQ
 
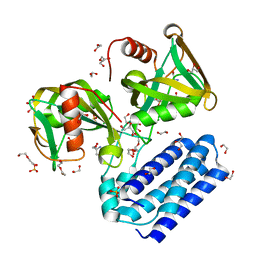 | | Crystal structure of UDP-X diphosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-10-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
2MRY
 
 | |
2MVB
 
 | |
4DE8
 
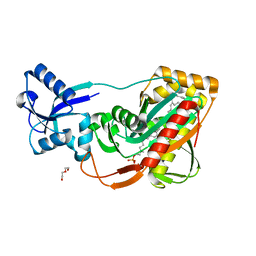 | | LytR-Cps2a-Psr family protein with bound octaprenyl monophosphate lipid | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl dihydrogen phosphate, Cps2A, DI(HYDROXYETHYL)ETHER | | Authors: | Eberhardt, A, Hoyland, C.N, Vollmer, D.V, Bisle, S, Cleverley, R.M, Johnsborg, O, Havarstein, L.S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2012-01-20 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Attachment of Capsular Polysaccharide to the Cell Wall in Streptococcus pneumoniae.
Microb Drug Resist, 18, 2012
|
|
2JTC
 
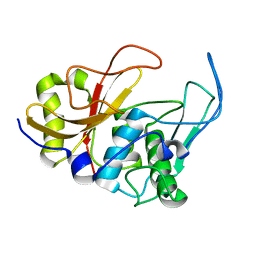 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
4EQB
 
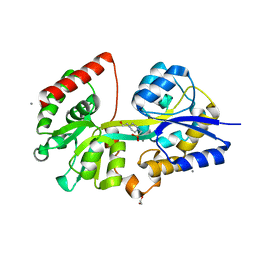 | | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein PotD from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES.
TO BE PUBLISHED
|
|
6MCB
 
 | |
4H1X
 
 | |
2L3A
 
 | | Solution NMR structure of homodimer protein SP_0782 (7-79) from Streptococcus pneumoniae Northeast Structural Genomics Consortium Target SpR104 . | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-10 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of hepothetical homodimer protein SP_0782 (7-79) from Streptococcus pneumoniae, Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
4H5G
 
 | | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 2
TO BE PUBLISHED
|
|
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
4E94
 
 | | Crystal structure of MccF-like protein from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, MccC family protein, SULFATE ION | | Authors: | Nocek, B, Tikhonov, A, Kwon, K, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of MccF-like protein from Streptococcus pneumoniae
TO BE PUBLISHED
|
|
4EM7
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 3-[3-(1H-pyrrolo[2,3-b]pyridin-5-yl)phenyl]propanoic acid, DNA topoisomerase IV, B subunit | | Authors: | Boriack-Sjodin, P.A, Manchester, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and Topoisomerase IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EMV
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 5-{2-(ethylcarbamoyl)-4-[3-(trifluoromethyl)-1H-pyrazol-1-yl]-1H-pyrrolo[2,3-b]pyridin-5-yl}pyridine-3-carboxylic acid, DNA topoisomerase IV, B subunit | | Authors: | Boriack-Sjodin, P.A, Manchester, J, Hull, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a novel azaindole class of antibacterial agents targeting the ATPase domains of DNA gyrase and Topoisomerase IV.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6JYX
 
 | | Structure of CbpJ from Streptococcus Pneumoniae TIGR4 | | Descriptor: | CHOLINE ION, Choline binding protein J, DI(HYDROXYETHYL)ETHER | | Authors: | Xu, Q, Zhang, J.W, Li, Q, Jiang, Y.L. | | Deposit date: | 2019-04-29 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the choline-binding protein CbpJ from Streptococcus pneumoniae.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
4HMP
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
4HMO
 
 | | Crystal structure of streptococcus pneumoniae TIGR4 PiaA in complex with Bis-tris propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Iron-compound ABC transporter, ... | | Authors: | Cheng, W, Li, Q, Jiang, Y.-L, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2012-10-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Streptococcus pneumoniae PiaA and Its Complex with Ferrichrome Reveal Insights into the Substrate Binding and Release of High Affinity Iron Transporters
Plos One, 8, 2013
|
|
2ND4
 
 | |
6N7F
 
 | | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD. | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Shabalin, I.G, Grabowski, M, Olphie, A, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.
To Be Published
|
|
2NOV
 
 | | Breakage-reunion domain of S.pneumoniae topo IV: crystal structure of a gram-positive quinolone target | | Descriptor: | DNA topoisomerase 4 subunit A | | Authors: | Laponogov, I, Veselkov, D.A, Sohi, M.K, Pan, X.S, Achari, A, Yang, C, Ferrara, J.D, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Breakage-Reunion Domain of Streptococcus pneumoniae Topoisomerase IV: Crystal Structure of a Gram-Positive Quinolone Target.
PLoS ONE, 2, 2007
|
|
4G41
 
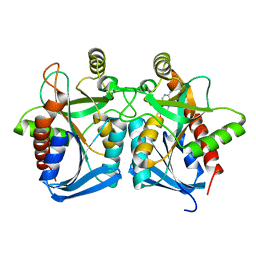 | | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Ponniah, K, Norris, G.E, Anderson, B.F, Brown, R.L, Tyler, P.C, Evans, G.B, Frohlich, R. | | Deposit date: | 2012-07-15 | | Release date: | 2012-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of s-adenosylhomocysteine nucleosidase from streptococcus pyogenes in complex with 5-methylthiotubericidin;
TO BE PUBLISHED
|
|
