4HPU
 
 | | Crystal structure of the catalytic subunit of cAMP-dependent protein kinase displaying partial phosphoryl transfer of AMP-PNP onto a substrate peptide | | Descriptor: | MAGNESIUM ION, MYRISTIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Bastidas, A.C, Steichen, J.M, Wu, J, Taylor, S.S. | | Deposit date: | 2012-10-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Phosphoryl transfer by protein kinase a is captured in a crystal lattice.
J.Am.Chem.Soc., 135, 2013
|
|
5VYW
 
 | | Crystal structure of Lactococcus lactis pyruvate carboxylase | | Descriptor: | BIOTIN, MANGANESE (II) ION, Pyruvate carboxylase | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6HVN
 
 | | CdaA-APO Y187A Mutant | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
6HVM
 
 | | Structural characterization of CdaA-APO | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
1ECJ
 
 | |
4XJ6
 
 | | Crystal structure of Escherichia coli DncV 3'-deoxy GTP bound form | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, VC0179-like protein | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
1EYK
 
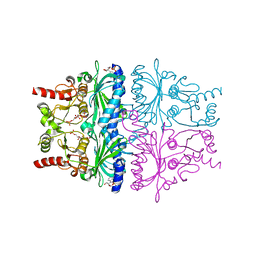 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH AMP, ZINC, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
1EYJ
 
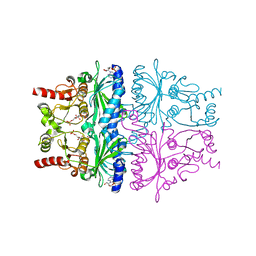 | | FRUCTOSE-1,6-BISPHOSPHATASE COMPLEX WITH AMP, MAGNESIUM, FRUCTOSE-6-PHOSPHATE AND PHOSPHATE (T-STATE) | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Choe, J, Honzatko, R.B. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of fructose 1,6-bisphosphatase: mechanism of catalysis and allosteric inhibition revealed in product complexes.
Biochemistry, 39, 2000
|
|
4QSL
 
 | |
3DAE
 
 | |
6V89
 
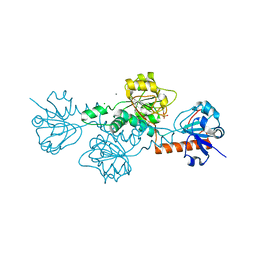 | | Human CtBP1 (28-375) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Royer, W.E. | | Deposit date: | 2019-12-10 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | NAD(H) phosphates mediate tetramer assembly of human C-terminal binding protein (CtBP).
J.Biol.Chem., 296, 2021
|
|
5WM2
 
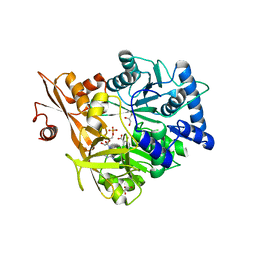 | |
5WM7
 
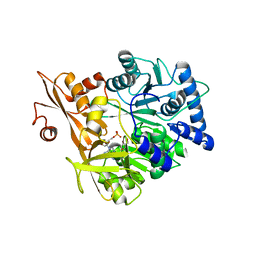 | | Crystal Structure of CahJ in Complex with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
3JXE
 
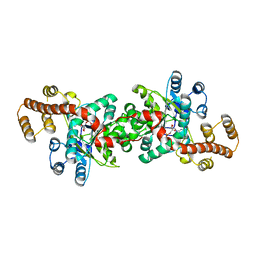 | | Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Yang, B, Ding, J. | | Deposit date: | 2009-09-19 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of P. horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase
To be Published
|
|
7RPW
 
 | |
7RPO
 
 | |
3TV1
 
 | | Crystal structure of RtcA.AMP product complex | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Chakravarty, A.K, Smith, P, Shuman, S. | | Deposit date: | 2011-09-19 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of RNA 3'-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
9NTK
 
 | | Chimeric Adenosine deaminase growth factor (ADGF) in complex with pentostatin monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Adenosine deaminase AGSA,Adenosine deaminase AGSA,Chimeric Adenosine deaminase growth factor, ... | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-18 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
9NTI
 
 | | Chimeric Adenosine deaminase growth factor (ADGF) apoenzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adenosine deaminase AGSA,Adenosine deaminase AGSA,Chimeric adenosine deaminase growth factor, ZINC ION | | Authors: | Kaur, G, Horton, J.R, Cheng, X. | | Deposit date: | 2025-03-18 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the substrate specificity of Helix pomatia AMP deaminase and a chimeric ADGF adenosine deaminase.
J.Biol.Chem., 2025
|
|
6VTE
 
 | | Naegleria gruberi RNA Ligase K170M mutant with AMP and Mn | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
3IDB
 
 | | Crystal structure of (108-268)RIIb:C holoenzyme of cAMP-dependent protein kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Brown, S.H.J, Wu, J, Kim, C, Alberto, K, Taylor, S.S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures.
J.Mol.Biol., 393, 2009
|
|
3IDC
 
 | | Crystal structure of (102-265)RIIb:C holoenzyme of cAMP-dependent protein kinase | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Brown, S.H.J, Wu, J, Kim, C, Alberto, K, Taylor, S.S. | | Deposit date: | 2009-07-20 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel isoform-specific interfaces revealed by PKA RIIbeta holoenzyme structures.
J.Mol.Biol., 393, 2009
|
|
1RZY
 
 | | Crystal structure of rabbit Hint complexed with N-ethylsulfamoyladenosine | | Descriptor: | 5'-O-(N-ETHYL-SULFAMOYL)ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Krakowiak, A.K, Pace, H.C, Blackburn, G.M, Adams, M, Mekhalfia, A, Kaczmarek, R, Baraniak, J, Stec, W.J, Brenner, C. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical, crystallographic, and mutagenic characterization of hint, the AMP-lysine hydrolase, with novel substrates and inhibitors
J.Biol.Chem., 279, 2004
|
|
6P63
 
 | |
6IJB
 
 | | Structure of 3-methylmercaptopropionate CoA ligase mutant K523A in complex with AMP and MMPA | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(methylsulfanyl)propanoic acid, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Shao, X, Cao, H.Y, Wang, P, Li, C.Y, Zhao, F, Peng, M, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
