7ABJ
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-1361 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-(3-(cyclopentyloxy)-4-methoxyphenyl)-2-isopropyl-4,4-dimethyl-2,4-dihydro-3H-pyrazol-3-one, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-1361
To be published
|
|
1E65
 
 | |
1E6N
 
 | | Chitinase B from Serratia marcescens inactive mutant E144Q in complex with N-acetylglucosamine-pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE B, GLYCEROL, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-21 | | Release date: | 2001-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7ABZ
 
 | | Structure of pre-accomodated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
1E7V
 
 | | Structure determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine | | Descriptor: | 2-MORPHOLIN-4-YL-7-PHENYL-4H-CHROMEN-4-ONE, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT | | Authors: | Walker, E.H, Pacold, M.E, Perisic, O, Stephens, L, Hawkins, P.T, Wymann, M.P, Williams, R.L. | | Deposit date: | 2000-09-08 | | Release date: | 2000-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine.
Mol.Cell, 6, 2000
|
|
1E8M
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN, MUTANT, COMPLEXED WITH INHIBITOR | | Descriptor: | GLYCEROL, N-[(benzyloxy)carbonyl]glycyl-L-proline, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2000-09-27 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Prolyl Oligopeptidase Substrate/ Inhibitor Complexes. Use of Inhibitor Binding for Titration of the Catalytic Histidine Residue
J.Biol.Chem., 276, 2001
|
|
1E9R
 
 | | Bacterial conjugative coupling protein TrwBdeltaN70. Trigonal form in complex with sulphate. | | Descriptor: | CONJUGAL TRANSFER PROTEIN TRWB, SULFATE ION | | Authors: | Gomis-Rueth, F.X, Moncalian, G, Cabezon, E, de la Cruz, F, Coll, M. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1EB2
 
 | | Trypsin inhibitor complex (BPO) | | Descriptor: | 3-[(Z)-AMINO(IMINO)METHYL]-N-[2-(4-BENZOYL-1-PIPERIDINYL)-2-OXO-1-PHENYLETHYL]BENZAMIDE, CALCIUM ION, SULFATE ION, ... | | Authors: | Wilkinson, K.W, Young, S.C, Liebeschuetz, J.W, Brady, R.L. | | Deposit date: | 2001-07-18 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pro_Select: Combining Structure-Based Drug Design and Array-Based Chemistry for Rapid Lead Discovery. 2. The Development of a Series of Highly Potent and Selective Factor Xa Inhibitors
J.Med.Chem., 45, 2002
|
|
1EBE
 
 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
7AAG
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-617 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-(4-methoxy-3-{4-[4-(1H-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy}phenyl)-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-04 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-617
To be published
|
|
7ABE
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-769 | | Descriptor: | 1,2-ETHANEDIOL, 1-cycloheptyl-3-[3-(cyclopentyloxy)-4-methoxyphenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-769
To be published
|
|
1ECY
 
 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
1ED6
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH L-NIO (H4B FREE) | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-01-26 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors.
Biochemistry, 40, 2001
|
|
7A1X
 
 | | KRASG12C GDP form in complex with Cpd1 | | Descriptor: | 3-(imidazol-1-ylmethyl)-1~{H}-indole, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mathieu, M, Steier, V. | | Deposit date: | 2020-08-14 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | KRAS G12C fragment screening renders new binding pockets.
Small Gtpases, 13, 2022
|
|
1EE3
 
 | | Cadmium-substituted bovine pancreatic carboxypeptidase A (alfa-form) at pH 7.5 and 2 mM chloride in monoclinic crystal form | | Descriptor: | CADMIUM ION, PROTEIN (CARBOXYPEPTIDASE A) | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-01-30 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1EEJ
 
 | | CRYSTAL STRUCTURE OF THE PROTEIN DISULFIDE BOND ISOMERASE, DSBC, FROM ESCHERICHIA COLI | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, THIOL:DISULFIDE INTERCHANGE PROTEIN | | Authors: | McCarthy, A.A, Haebel, P.W, Torronen, A, Rybin, V, Baker, E.N, Metcalf, P. | | Deposit date: | 2000-01-31 | | Release date: | 2000-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli.
Nat.Struct.Biol., 7, 2000
|
|
1EH8
 
 | |
1ELL
 
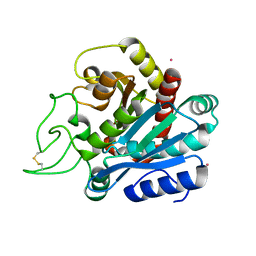 | | CADMIUM-SUBSTITUTED BOVINE PANCREATIC CARBOXYPEPTIDASE A (ALFA-FORM) AT PH 7.5 AND 0.25 M CHLORIDE IN MONOCLINIC CRYSTAL FORM. | | Descriptor: | CADMIUM ION, CARBOXYPEPTIDASE A | | Authors: | Jensen, F, Bukrinsky, T, Bjerrum, J, Larsen, S. | | Deposit date: | 2000-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Three high-resolution crystal structures of cadmium-substituted carboxypeptidase A provide insight into the enzymatic function
J.BIOL.INORG.CHEM., 7, 2002
|
|
1ENY
 
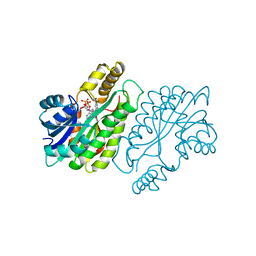 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
5HAU
 
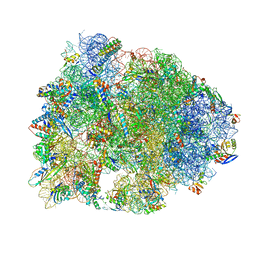 | | Crystal structure of antimicrobial peptide Bac7(1-19) bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Roy, R.N, Lomakin, I.B, Florin, T, Mankin, A.S, Steitz, T.A. | | Deposit date: | 2015-12-30 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of proline-rich peptides bound to the ribosome reveal a common mechanism of protein synthesis inhibition.
Nucleic Acids Res., 44, 2016
|
|
1ENZ
 
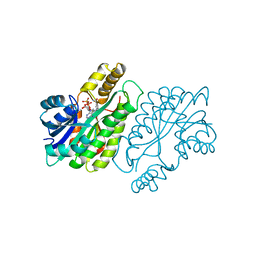 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
6ZPP
 
 | |
1E3V
 
 | | Crystal structure of ketosteroid isomerase from Psedomonas putida complexed with deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, STEROID DELTA-ISOMERASE | | Authors: | Ha, N.-C, Kim, M.-S, Kim, J.-S, Oh, B.-H. | | Deposit date: | 2000-06-24 | | Release date: | 2001-03-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Large Pka Perturbations of an Inhibitor and a Catalytic Group at an Enzyme Active Site, a Mechanistic Basis for Catalytic Power of Many Enzymes
J.Biol.Chem., 275, 2000
|
|
1DY4
 
 | | CBH1 IN COMPLEX WITH S-PROPRANOLOL | | Descriptor: | 1-(ISOPROPYLAMINO)-3-(1-NAPHTHYLOXY)-2-PROPANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Stahlberg, J, Henriksson, H, Divne, C, Isaksson, R, Pettersson, G, Johansson, G, Jones, T.A. | | Deposit date: | 2000-01-26 | | Release date: | 2000-12-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Enantiomer Binding and Separation of a Common Beta-Blocker: Crystal Structure of Cellobiohydrolase Cel7A with Bound (S)-Propranolol at 1.9 A Resolution
J.Mol.Biol., 305, 2001
|
|
1DTS
 
 | | CRYSTAL STRUCTURE OF AN ATP DEPENDENT CARBOXYLASE, DETHIOBIOTIN SYNTHASE, AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-03-28 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of an ATP-dependent carboxylase, dethiobiotin synthetase, at 1.65 A resolution.
Structure, 2, 1994
|
|
