4PPQ
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-CR | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
7W5L
 
 | | The crystal structure of the oxidized form of Gluconobacter oxydans WSH-004 SNDH | | Descriptor: | L-sorbosone dehydrogenase, NAD(P) dependent | | Authors: | Li, D, Hou, X.D, Rao, Y.J, Zhou, J.W, Chen, J. | | Deposit date: | 2021-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into the Catalytic Mechanisms of an L-Sorbosone Dehydrogenase.
Adv Sci, 10, 2023
|
|
7OYD
 
 | | Cryo-EM structure of a rabbit 80S ribosome with zebrafish Dap1b | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
2VGR
 
 | |
6JGO
 
 | | Crystal structure of barley exohydrolaseI W434H mutant in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
3E74
 
 | |
4PQY
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-N4 | | Descriptor: | GLYCEROL, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
6DKT
 
 | |
4PCP
 
 | | Crystal structure of Phosphotriesterase variant R0 | | Descriptor: | CACODYLATE ION, Phosphotriesterase variant PTE-R0, ZINC ION | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-04-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
5WQN
 
 | |
7W32
 
 | | Deactive state CI from DQ-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4PRL
 
 | |
2VIG
 
 | | Crystal structure of human short-chain acyl CoA dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A PERSULFIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pike, A.C.W, Pantic, N, Parizotto, E, Gileadi, O, Ugochukwu, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Oppermann, U. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Short-Chain Acyl Coa Dehydrogenase
To be Published
|
|
3HWL
 
 | | Crystal Structure of T4 lysozyme with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 131 | | Descriptor: | AZIDE ION, CHLORIDE ION, Lysozyme | | Authors: | Fleissner, M.R, Cascio, D, Schultz, P.G, Hubbell, W.L. | | Deposit date: | 2009-06-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed spin labeling of a genetically encoded unnatural amino acid.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7P2V
 
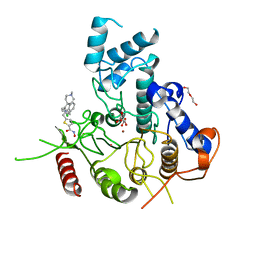 | | Crystal structure of Schistosoma mansoni HDAC8 in complex with a 4-chlorophenyl-spiroindoline capped hydroxamate-based inhibitor, bound to a novel site | | Descriptor: | 5-[[(2S)-2-(4-chlorophenyl)-1'-methyl-spiro[2H-indole-3,4'-piperidine]-1-yl]methyl]-N-oxidanyl-thiophene-2-carboxamide, DI(HYDROXYETHYL)ETHER, Histone deacetylase 8, ... | | Authors: | Saccoccia, F, Gemma, S, Campiani, G, Ruberti, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Schistosoma mansoni histone deacetylase 8 reveal a novel binding site for allosteric inhibitors.
J.Biol.Chem., 298, 2022
|
|
6JKG
 
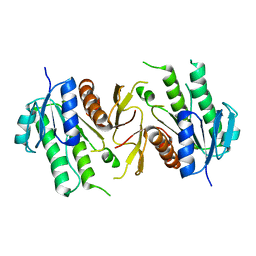 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
3EBU
 
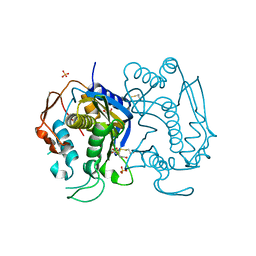 | | Replacement of Val3 in Human Thymidylate Synthase Affects Its Kinetic Properties and Intracellular Stability | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Thymidylate synthase | | Authors: | Huang, X, Gibson, L.M, Bell, B.J, Lovelace, L.L, Pena, M.M, Berger, F.G, Berger, S.H. | | Deposit date: | 2008-08-28 | | Release date: | 2010-03-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Replacement of Val3 in human thymidylate synthase affects its kinetic properties and intracellular stability .
Biochemistry, 49, 2010
|
|
7W4K
 
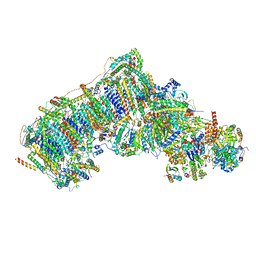 | | Deactive state CI from Q1-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4PSP
 
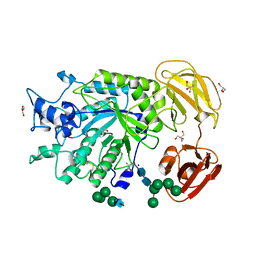 | | Crystal Structure of GH29 family alpha-L-fucosidase from Fusarium graminearum in the open form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-fucosidase GH29, ... | | Authors: | Cao, H, Walton, J, Brumm, P, Phillips Jr, G.N. | | Deposit date: | 2014-03-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Structure and Substrate Specificity of a Eukaryotic Fucosidase from Fusarium graminearum.
J.Biol.Chem., 289, 2014
|
|
7W1T
 
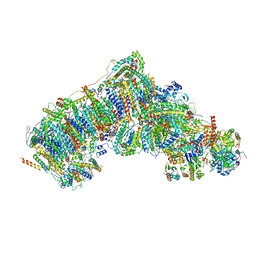 | | Active state CI from Rotenone dataset, Subclass 1 | | Descriptor: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-20 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W2L
 
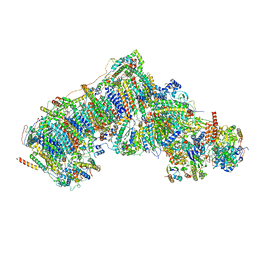 | | Deactive state CI from Rotenone-NADH dataset, Subclass 2 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J.K, Yang, M.J. | | Deposit date: | 2021-11-24 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4PUF
 
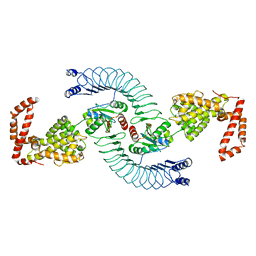 | | Complex between the Salmonella T3SS effector SlrP and its human target thioredoxin-1 | | Descriptor: | E3 ubiquitin-protein ligase SlrP, Thioredoxin | | Authors: | Zouhir, S, Bernal-Bayard, J, Cordero-Alba, M, Cardenal-Munoz, E, Guimaraes, B, Lazar, N, Ramos-Morales, F, Nessler, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | The structure of the Slrp-Trx1 complex sheds light on the autoinhibition mechanism of the type III secretion system effectors of the NEL family.
Biochem.J., 464, 2014
|
|
3E9B
 
 | |
2V7P
 
 | | Crystal structure of lactate dehydrogenase from Thermus Thermophilus HB8 (Holo form) | | Descriptor: | L-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID | | Authors: | Coquelle, N, Fioravanti, E, Weik, M, Vellieux, F. | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environments.
J. Mol. Biol., 374, 2007
|
|
4N2Z
 
 | | Crystal Structure of the alpha-L-arabinofuranosidase PaAbf62A from Podospora anserina in complex with cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
