2H2I
 
 | | The Structural basis of Sirtuin Substrate Affinity | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | Authors: | Cosgrove, M.S, Wolberger, C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
2H2D
 
 | |
2H2F
 
 | |
2KA5
 
 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2H2H
 
 | |
6MAX
 
 | |
1LW4
 
 | |
1LW5
 
 | |
1M6S
 
 | | Crystal Structure Of Threonine Aldolase | | Descriptor: | CALCIUM ION, CHLORIDE ION, L-allo-threonine aldolase | | Authors: | Burley, S.K, Kielkopf, C.L. | | Deposit date: | 2002-07-17 | | Release date: | 2002-12-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of Threonine Aldolase Complexes: Structural Basis of Substrate Recognition
Biochemistry, 41, 2002
|
|
2H2G
 
 | |
3CWO
 
 | | A beta/alpha-barrel built by the combination of fragments from different folds | | Descriptor: | SULFATE ION, beta/alpha-barrel protein based on 1THF and 1TMY | | Authors: | Bharat, T.A.M, Eisenbeis, S, Zeth, K, Hocker, B. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A beta alpha-barrel built by the combination of fragments from different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5JCP
 
 | | RhoGAP domain of ARAP3 in complex with RhoA in the transition state | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3,Linker,Transforming protein RhoA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bao, H, Li, F, Wang, C, Wang, N, Jiang, Y, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
6DTR
 
 | | Apo T. maritima MalE3 | | Descriptor: | SULFATE ION, maltose-binding protein MalE3 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTS
 
 | | Maltotetraose bound T. maritima MalE2 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTT
 
 | | Apo T. maritima MalE2 | | Descriptor: | maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | Authors: | Cuneo, M.J, Shukla, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
5WSF
 
 | | Crystal structure of a cupin protein (tm1459) in osmium (Os)-substituted form II | | Descriptor: | OSMIUM ION, Uncharacterized protein tm1459 | | Authors: | Fujieda, N, Nakano, T, Taniguchi, Y, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | A Well-Defined Osmium-Cupin Complex: Hyperstable Artificial Osmium Peroxygenase
J. Am. Chem. Soc., 2017
|
|
5TDY
 
 | | Structure of cofolded FliFc:FliGn complex from Thermotoga maritima | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG | | Authors: | Lynch, M.J, Levenson, R, Kim, E.A, Sircar, R, Blair, D.F, Dahlquist, F.W, Crane, B.R. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Co-Folding of a FliF-FliG Split Domain Forms the Basis of the MS:C Ring Interface within the Bacterial Flagellar Motor.
Structure, 25, 2017
|
|
3VRS
 
 | | Crystal structure of fluoride riboswitch, soaked in Mn2+ | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MANGANESE (II) ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
6Q44
 
 | | Est3 telomerase subunit in the yeast Hansenula polymorpha | | Descriptor: | Uncharacterized protein | | Authors: | Mantsyzov, A.B, Mariasina, S.S, Petrova, O.A, Efimov, S.V, Dontsova, O.A, Polshakov, V.I. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Sci Rep, 10, 2020
|
|
4W9M
 
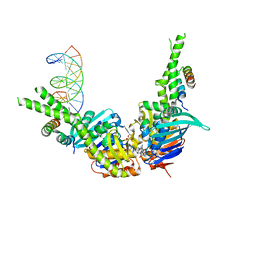 | | AMPPNP bound Rad50 in complex with dsDNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*CP*AP*CP*CP*GP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*CP*GP*GP*TP*GP*AP*CP*CP*GP*AP*CP*C)-3'), Exonuclease, ... | | Authors: | Rojowska, A, Lammens, K. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Rad50 DNA double-strand break repair protein in complex with DNA.
Embo J., 33, 2014
|
|
7NQD
 
 | |
7NQF
 
 | |
7NQE
 
 | |
7P9J
 
 | |
