8U6J
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(5-chloro-2-((2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ746), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-[2-(5-chloro-2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8OJT
 
 | | Crystal structure of the human IgD Fab - structure Fab2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, ... | | Authors: | Davies, A.M, Beavil, R.L, McDonnell, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the human IgD Fab reveal insights into C H 1 domain diversity.
Mol.Immunol., 159, 2023
|
|
8F8M
 
 | | LRH-1 bound to small molecule Tet and fragment of coactivator Tif2 | | Descriptor: | (1~{R},3~{a}~{R},6~{a}~{R})-4-phenyl-3~{a}-(1-phenylethenyl)-5-[9-(1~{H}-1,2,3,4-tetrazol-5-yl)nonyl]-2,3,6,6~{a}-tetrahydro-1~{H}-pentalen-1-ol, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Cato, M.L, Ortlund, E.A. | | Deposit date: | 2022-11-22 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Isosteric improvements to liver receptor homolog-1 small molecule modulators
To Be Published
|
|
6VQS
 
 | |
6X1Q
 
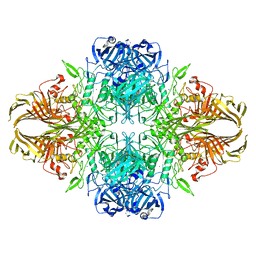 | | 1.8 Angstrom resolution structure of b-galactosidase with a 200 kV cryoARM electron microscope | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, SODIUM ION | | Authors: | Merk, A, Fukumura, T, Zhu, X, Darling, J, Grisshammer, R, Ognjenovic, J, Subramaniam, S. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.8 Å) | | Cite: | 1.8 angstrom resolution structure of beta-galactosidase with a 200 kV CRYO ARM electron microscope.
Iucrj, 7, 2020
|
|
6X1U
 
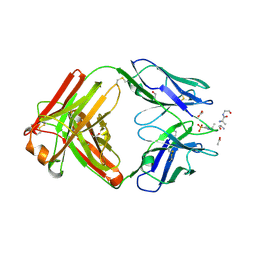 | | Structure of pHis Fab (SC39-4) in complex with pHis mimetic peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACLYana-3-pTza peptide, ... | | Authors: | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9BCZ
 
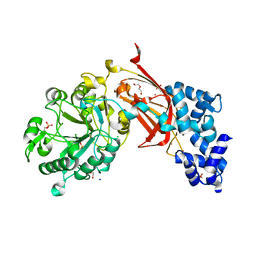 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
8U6E
 
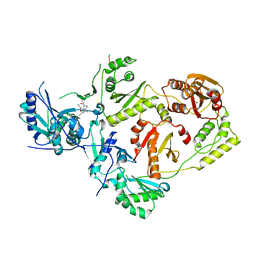 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenethyl)-N-methylacrylamide (JLJ738), a non-nucleoside inhibitor | | Descriptor: | MAGNESIUM ION, N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenyl]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
7TQU
 
 | |
6W4C
 
 | | Crystal structure of HAO1 in complex with indazole acid inhibitor - compound 5 | | Descriptor: | 5-[[3-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-2-oxidanyl-phenyl]methylamino]-2~{H}-indazole-3-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
6W45
 
 | | Crystal structure of HAO1 in complex with biaryl acid inhibitor - compound 3 | | Descriptor: | 2-chloranyl-4-[2-[[(6-chloranyl-1~{H}-indol-2-yl)carbonyl-methyl-amino]methyl]-5-fluoranyl-phenyl]benzoic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Ferguson, A.D. | | Deposit date: | 2020-03-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Novel, Potent Inhibitors of Hydroxy Acid Oxidase 1 (HAO1) Using DNA-Encoded Chemical Library Screening.
J.Med.Chem., 64, 2021
|
|
8G3T
 
 | | MBP-Mcl1 in complex with ligand 12 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3U
 
 | | MBP-Mcl1 in complex with ligand 21 | | Descriptor: | (1'S,3aS,5R,16R,17S,19Z,21R,21aR)-6'-chloro-20-fluoro-21-{[(5S,9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]methyl}-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-(ethanediylidene)-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, 1,2-ETHANEDIOL, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
8G3S
 
 | | MBP-Mcl1 in complex with ligand 11 | | Descriptor: | (1'S,3aS,5R,16R,17S,19E,21S,21aR)-6'-chloro-21-methoxy-16,17-dimethyl-2,3,3',3a,4',16,17,18,21,21a-decahydro-2'H,6H,8H-15lambda~6~-spiro[10,12-etheno-15lambda~6~-furo[3,2-i][1,4]oxazepino[3,4-f][1,2,7]thiadiazacyclohexadecine-7,1'-naphthalene]-13,15,15(4H,14H)-trione, FORMIC ACID, Maltodextrin-binding protein, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of an Oral, Beyond-Rule-of-Five Mcl-1 Protein-Protein Interaction Modulator with the Potential of Treating Hematological Malignancies.
J.Med.Chem., 66, 2023
|
|
9LSF
 
 | | Crystal structure of mRFP1 with a grafted calcium-binding sequence and one bound calcium ion in a calcium-free solution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uehara, R, Kamiya, Y, Maeda, S, Okamoto, K, Toya, S, Chiba, R, Amesaka, H, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enhanced secretion through type 1 secretion system by grafting a calcium-binding sequence to modify the folding of cargo proteins.
Protein Sci., 34, 2025
|
|
9LSA
 
 | | Crystal structure of mRFP1 with a grafted calcium-binding sequence and one bound calcium ion in a calcium-containing solution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Red fluorescent protein,grafted calcium-binding sequence, ... | | Authors: | Uehara, R, Kamiya, Y, Maeda, S, Okamoto, K, Toya, S, Chiba, R, Amesaka, H, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enhanced secretion through type 1 secretion system by grafting a calcium-binding sequence to modify the folding of cargo proteins.
Protein Sci., 34, 2025
|
|
8PWE
 
 | | Crystal structure of VDR complex with Novel Des-C-Ring and Aromatic-D-Ring analog 3a | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-hept-3-ynyl)phenyl]pent-2-enylidene]cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-07-20 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Further Studies on the Highly Active Des-C-Ring and Aromatic-D-Ring Analogues of 1 alpha ,25-Dihydroxyvitamin D 3 (Calcitriol): Refinement of the Side Chain.
J.Med.Chem., 66, 2023
|
|
6O3D
 
 | |
6HMB
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 Gene product) IN COMPLEX WITH the inhibitor CX-4945 (Silmitasertib) | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6OL7
 
 | |
6STU
 
 | | Adenovirus 30 Fiber Knob protein | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLUTAMIC ACID | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Broad sialic acid usage amongst species D human adenovirus
npj Viruses, 1, 2023
|
|
9NSY
 
 | | OXA-23-NA-1-157, 4 minute soak | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase OXA-23, SULFATE ION | | Authors: | Smith, C.A, Toth, M, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-03-17 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dual mechanism of the OXA-23 carbapenemase inhibition by the carbapenem NA-1-157.
Antimicrob.Agents Chemother., 69, 2025
|
|
9NSX
 
 | | OXA-23-NA-1-157, 3 minute soak | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase OXA-23, SULFATE ION | | Authors: | Smith, C.A, Toth, M, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-03-17 | | Release date: | 2025-08-06 | | Last modified: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Dual mechanism of the OXA-23 carbapenemase inhibition by the carbapenem NA-1-157.
Antimicrob.Agents Chemother., 69, 2025
|
|
8TD3
 
 | |
8TDD
 
 | | Structure of PYCR1 complexed with NADH and 2-(furan-2-yl)acetic acid | | Descriptor: | (furan-2-yl)acetic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
