1U0E
 
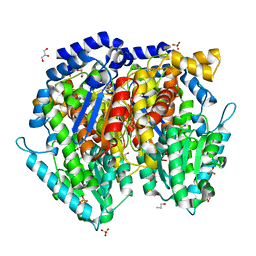 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1ZXY
 
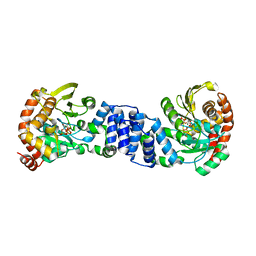 | | Anthranilate Phosphoribosyltransferase from Sulfolobus solfataricus in complex with PRPP and Magnesium | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Marino, M, Deuss, M, Sterner, R, Mayans, O. | | Deposit date: | 2005-06-09 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
J.Biol.Chem., 281, 2006
|
|
5X8L
 
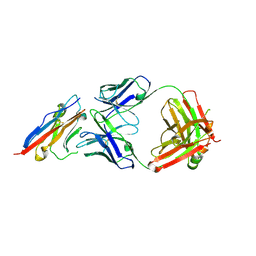 | | PD-L1 in complex with atezolizumab | | Descriptor: | Programmed cell death 1 ligand 1, atezolizumab heavy chain, atezolizumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
1H4K
 
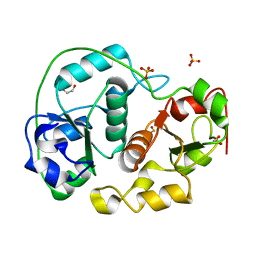 | | Sulfurtransferase from Azotobacter vinelandii in complex with hypophosphite | | Descriptor: | 1,2-ETHANEDIOL, HYPOPHOSPHITE, SULFATE ION, ... | | Authors: | Bordo, D, Forlani, F, Spallarossa, A, Colnaghi, R, Carpen, A, Pagani, S, Bolognesi, M. | | Deposit date: | 2001-05-11 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Persulfurated Cysteine Promotes Active Site Reactivity in Azotobacter Vinelandii Rhodanse
Biol.Chem., 382, 2001
|
|
4OLF
 
 | |
5XAU
 
 | | Crystal structure of integrin binding fragment of laminin-511 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Takizawa, M, Arimori, T, Kitago, Y, Takagi, J, Sekiguchi, K. | | Deposit date: | 2017-03-15 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic basis for the recognition of laminin-511 by alpha 6 beta 1 integrin.
Sci Adv, 3, 2017
|
|
2PA5
 
 | | Crystal structure of human protein tyrosine phosphatase PTPN9 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THIOCYANATE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Pike, A.C.W, Savitsky, P, Papagrigoriou, E, Turnbull, A, Uppenberg, J, Bunkoczi, G, Salah, E, Das, S, von Delft, F, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2PCY
 
 | |
2A2N
 
 | | Crystal Structure of the peptidylprolyl isomerase domain of Human PPWD1 | | Descriptor: | GLYCEROL, peptidylprolyl isomerase domain and WD repeat containing 1 | | Authors: | Walker, J.R, Davis, T.L, Newman, E.M, Mackenzie, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-22 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of human WD40 repeat-containing peptidylprolyl isomerase (PPWD1).
Febs J., 275, 2008
|
|
3QK8
 
 | |
1H96
 
 | | recombinant mouse L-chain ferritin | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, SULFATE ION | | Authors: | Granier, T, Gallois, B, D'Estaintot, B.L, Dautant, A, Chevalier, J.M, Mellado, J.M, Beaumont, C, Santambrogio, P, Arosio, P, Precigoux, G. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Mouse L-Chain Ferritin at 1.6 A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1H62
 
 | | Structure of Pentaerythritol tetranitrate reductase in complex with 1,4-androstadien-3,17-dione | | Descriptor: | ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE | | Authors: | Barna, T.M, Moody, P.C.E. | | Deposit date: | 2001-06-04 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Pentaerythritol Tetranitrate Reductase: "Flipped" Binding Geometries for Steroid Substrates in Different Redox States of the Enzyme
J.Mol.Biol., 310, 2001
|
|
2OPB
 
 | | Structure of K57A hPNMT with inhibitor 3-fluoromethyl-7-thiomorpholinosulfonamide-THIQ and AdoHcy | | Descriptor: | (3R)-3-(FLUOROMETHYL)-7-(THIOMORPHOLIN-4-YLSULFONYL)-1,2,3,4-TETRAHYDROISOQUINOLINE, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase.
J.Med.Chem., 50, 2007
|
|
7EH5
 
 | | Cryo-EM structure of SARS-CoV-2 S-D614G variant in complex with neutralizing antibodies, RBD-chAb15 and RBD-chAb45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RBD-chAb15, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-03-28 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4G0W
 
 | | Human topoisomerase iibeta in complex with DNA and ametantrone | | Descriptor: | 1,4-bis({2-[(2-hydroxyethyl)amino]ethyl}amino)anthracene-9,10-dione, DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Wu, C.C, Li, T.K, Li, Y.C, Chan, N.L. | | Deposit date: | 2012-07-10 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs
Nucleic Acids Res., 41, 2013
|
|
4C0M
 
 | | Crystal Structure of the N terminal domain of wild type TRIF (TIR- domain-containing adapter-inducing interferon-beta) | | Descriptor: | TIR DOMAIN-CONTAINING ADAPTER MOLECULE 1 | | Authors: | Ullah, M.O, Ve, T, Mangan, M, Alaidarous, M, Sweet, M.J, Mansell, A, Kobe, B. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Tlr Signalling Adaptor Trif/Ticam-1 Has an N-Terminal Helical Domain with Structural Similarity to Ifit Proteins
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4C1C
 
 | | Crystal structure of the metallo-beta-lactamase BCII with D-captopril | | Descriptor: | 1-(3-MERCAPTO-2-METHYL-PROPIONYL)-PYRROLIDINE-2-CARBOXYLIC ACID, BETA-LACTAMASE 2, GLYCEROL, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
6KK6
 
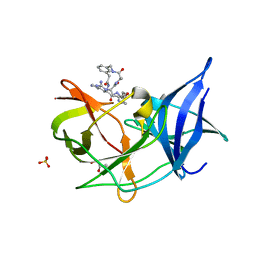 | | Crystal structure of Zika NS2B-NS3 protease with compound 16 | | Descriptor: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-5-methyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, GLYCEROL, NS3 protease, ... | | Authors: | Quek, J.P. | | Deposit date: | 2019-07-23 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
3E8M
 
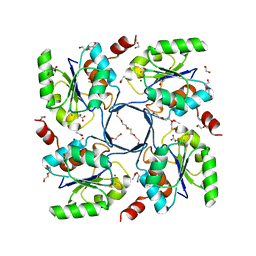 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acylneuraminate cytidylyltransferase, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
3Q84
 
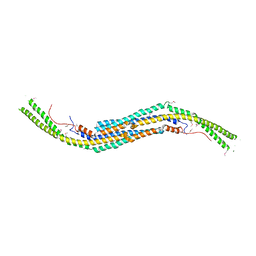 | | Crystal structure of human PACSIN 1 F-BAR domain | | Descriptor: | CALCIUM ION, Protein kinase C and casein kinase substrate in neurons protein 1 | | Authors: | Bai, X. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human PACSIN 1 F-BAR domain
To be Published
|
|
5E8A
 
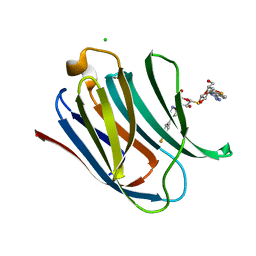 | | Crystal structure of Human galectin-3 CRD in complex with 4-fluophenyl-1,2,3-triazolyl thiodigalactoside inhibitor | | Descriptor: | 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2015-10-13 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galectin-3-Binding Glycomimetics that Strongly Reduce Bleomycin-Induced Lung Fibrosis and Modulate Intracellular Glycan Recognition.
Chembiochem, 17, 2016
|
|
1HGT
 
 | |
4O7C
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-614067-R | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5Z)-5-(1-nitroso-2,3-dihydro-5H-inden-5-ylidene)-2-(piperidin-4-yl)-3,5-dihydro-4H-imidazol-4-ylidene]-1,4-dihydropyridine, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
4O9D
 
 | | Structure of Dos1 propeller | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Rik1-associated factor 1 | | Authors: | Kuscu, C, Schalch, T, Joshua-Tor, L. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRL4-like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ZO8
 
 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with sophorose | | Descriptor: | Lin1840 protein, MAGNESIUM ION, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
