8E4W
 
 | |
3FTX
 
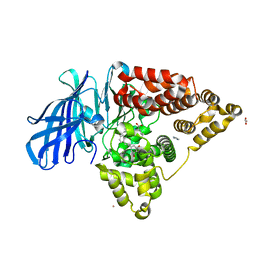 | | Leukotriene A4 hydrolase in complex with dihydroresveratrol and bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 5-[2-(4-hydroxyphenyl)ethyl]benzene-1,3-diol, ACETATE ION, ... | | Authors: | Davies, D.R. | | Deposit date: | 2009-01-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of leukotriene A4 hydrolase inhibitors using metabolomics biased fragment crystallography.
J.Med.Chem., 52, 2009
|
|
3FU0
 
 | |
3FUH
 
 | |
1GUQ
 
 | | STRUCTURE OF NUCLEOTIDYLTRANSFERASE COMPLEXED WITH UDP-GLUCOSE | | Descriptor: | FE (III) ION, GALACTOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, POTASSIUM ION, ... | | Authors: | Thoden, J.B, Rayment, I, Holden, H. | | Deposit date: | 1996-10-23 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the H166G site-directed mutant of galactose-1-phosphate uridylyltransferase complexed with either UDP-glucose or UDP-galactose: detailed description of the nucleotide sugar binding site.
Biochemistry, 36, 1997
|
|
1GUP
 
 | | STRUCTURE OF NUCLEOTIDYLTRANSFERASE COMPLEXED WITH UDP-GALACTOSE | | Descriptor: | FE (III) ION, GALACTOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Rayment, I, Holden, H. | | Deposit date: | 1996-10-23 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the H166G site-directed mutant of galactose-1-phosphate uridylyltransferase complexed with either UDP-glucose or UDP-galactose: detailed description of the nucleotide sugar binding site.
Biochemistry, 36, 1997
|
|
1GOL
 
 | |
5ACU
 
 | | VIM-2-NAT, Discovery of novel inhibitor scaffolds against the metallo- beta-lactamase VIM-2 by SPR based fragment screening | | Descriptor: | BETA-LACTAMASE, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Christopeit, T, Carlsen, T.J.O, Helland, R, Leiros, H.K.S. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel Inhibitor Scaffolds Against the Metallo-Beta-Lactamase Vim-2 by Spr Based Fragment Screening
J.Med.Chem., 58, 2015
|
|
7RDQ
 
 | |
8QCI
 
 | | FCGBP D10 Assembly Segment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Yeshaya, N, Fass, D. | | Deposit date: | 2023-08-27 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | VWD domain stabilization by autocatalytic Asp-Pro cleavage.
Protein Sci., 33, 2024
|
|
4G6I
 
 | | Crystallographic structure of trimeric riboflavin synthase from Brucella abortus in complex with roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, Riboflavin synthase subunit alpha | | Authors: | Serer, M.I, Bonomi, H.R, Guimaraes, B.G, Rossi, R.C, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2012-07-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystallographic and kinetic study of riboflavin synthase from Brucella abortus, a chemotherapeutic target with an enhanced intrinsic flexibility.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6WW1
 
 | | Crystal structure of the LmFPPS mutant E97Y | | Descriptor: | 3-butyl-1-(2,2-diphosphonoethyl)pyridinium, ACETATE ION, CALCIUM ION, ... | | Authors: | Maheshwari, S, Kim, Y.S, Gabelli, S.B. | | Deposit date: | 2020-05-07 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying Structural Determinants of Product Specificity in Leishmania major Farnesyl Diphosphate Synthase.
Biochemistry, 59, 2020
|
|
3H9A
 
 | | Crystal structure of BacB, an enzyme involved in Bacilysin synthesis, in triclinic form | | Descriptor: | 3-PHENYLPYRUVIC ACID, Bacilysin biosynthesis protein bacB, COBALT (II) ION, ... | | Authors: | Rajavel, M, Gopal, B. | | Deposit date: | 2009-04-30 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Analysis of multiple crystal forms of Bacillus subtilis BacB suggests a role for a metal ion as a nucleant for crystallization
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2BG8
 
 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH4.5 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT and 1mM TCEP-HCl were used as reducing agents. | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, SULFATE ION, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
1I1Z
 
 | | MUTANT HUMAN LYSOZYME (Q86D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I4H
 
 | | Crystal structure of Zn2+ soaked Staphylococcal enterotoxin A mutant H187A | | Descriptor: | ENTEROTOXIN TYPE A, ZINC ION | | Authors: | Hakansson, M, Antonsson, P, Bjork, P, Svensson, L.A. | | Deposit date: | 2001-02-21 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cooperative zinc binding in a staphylococcal enterotoxin A mutant mimics the SEA-MHC class II interaction
J.Biol.Inorg.Chem., 6, 2001
|
|
1I20
 
 | | MUTANT HUMAN LYSOZYME (A92D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
5NIG
 
 | |
1I22
 
 | | MUTANT HUMAN LYSOZYME (A83K/Q86D/A92D) | | Descriptor: | CALCIUM ION, LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I4G
 
 | | Crystal structure of Staphylococcal enterotoxin A mutant H187A with reduced Zn2+ affinity | | Descriptor: | ENTEROTOXIN TYPE A, SULFATE ION, ZINC ION | | Authors: | Hakansson, M, Antonsson, P, Bjork, P, Svensson, L.A. | | Deposit date: | 2001-02-21 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative zinc binding in a staphylococcal enterotoxin A mutant mimics the SEA-MHC class II interaction
J.Biol.Inorg.Chem., 6, 2001
|
|
1DZE
 
 | | Structure of the M Intermediate of Bacteriorhodopsin trapped at 100K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Iizuka, T, Kamiya, N, Adachi, S, Kouyama, T. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the M Intermediate of Bacteriorhodopsin: Allosteric Structural Changes Mediated by Sliding Movement of a Transmembrane Helix
J.Mol.Biol., 341, 2004
|
|
5UIN
 
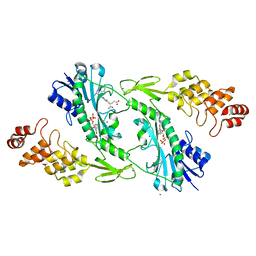 | | X-ray structure of the W305A variant of the FdtF N-formyltransferase from salmonella enteric O60 | | Descriptor: | CHLORIDE ION, Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
1ICJ
 
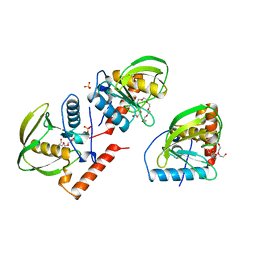 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1ZPR
 
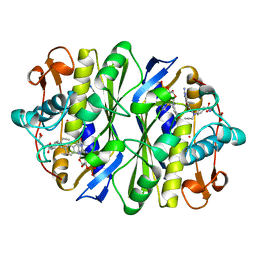 | | E. COLI THYMIDYLATE SYNTHASE MUTANT E58Q IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Stout, T.J, Rutenber, E.E, Stroud, R.M. | | Deposit date: | 1996-10-15 | | Release date: | 1997-07-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An essential role for water in an enzyme reaction mechanism: the crystal structure of the thymidylate synthase mutant E58Q.
Biochemistry, 35, 1996
|
|
1I6S
 
 | | T4 LYSOZYME MUTANT C54T/C97A/N101A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Kovall, R.A, Baldwin, E.P, Matthews, B.W. | | Deposit date: | 2001-03-04 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
