2UUO
 
 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-{[6-(PENTYLOXY)NAPHTHALEN-2-YL]SULFONYL}-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
3WX9
 
 | | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PMP, GLA, 4AD, 2OG, GLU and KYA | | Descriptor: | (2E)-pent-2-enedioic acid, 2-OXOGLUTARIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Okada, K, Angkawidjaja, C, Koga, Y, Kanaya, S. | | Deposit date: | 2014-07-28 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of Pyrococcus horikoshii kynurenine aminotransferase in complex with PMP, GLA, 4AD, 2OG, GLU and KYA
To be Published
|
|
2N8F
 
 | |
7VFU
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to ziconotide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7PZ9
 
 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
6YAA
 
 | | Structure of the (SR) Ca2+-ATPase bound to the inhibitor compound CAD204520 and TNP-ATP | | Descriptor: | 4-[2-[(2~{R})-2-[3-propyl-6-(trifluoromethyloxy)-1~{H}-indol-2-yl]piperidin-1-yl]ethyl]morpholine, POTASSIUM ION, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, ... | | Authors: | Heit, S, Marchesini, M, Gherli, A, Montanaro, A, Patrizi, L, Sorrentino, C, Pagliaro, L, Rompietti, C, Kitara, S, Olesen, C.E, Moller, J.V, Savi, M, Bocchi, L, Vilella, R, Rizzi, F, Baglione, M, Rastelli, G, Loiacona, C, La Starza, R, Mecucci, C, Stegmair, K, Aversa, F, Stilli, D, Lund Winther, A.M, Sportoletti, P, Dalby-Brown, W, Roti, G, Bublitz, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Cell Chem Biol, 27, 2020
|
|
2UUP
 
 | | Crystal structure of MurD ligase in complex with D-Glu containing sulfonamide inhibitor | | Descriptor: | N-({6-[(4-CYANOBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)-D-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | Authors: | Humljan, J, Kotnik, M, Contreras-Martel, C, Blanot, D, Urleb, U, Dessen, A, Solmajer, T, Gobec, S. | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Novel naphthalene-N-sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a key peptidoglycan biosynthesis enzyme.
J. Med. Chem., 51, 2008
|
|
7OI0
 
 | | E.coli delta rbfA pre-30S ribosomal subunit class D | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Maksimova, E, Korepanov, A, Baymukhametov, T, Kravchenko, O, Stolboushkina, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center.
Int J Mol Sci, 22, 2021
|
|
6AHW
 
 | |
5ZYO
 
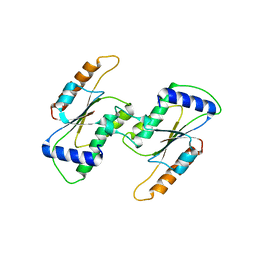 | |
7R6H
 
 | | RHCC in complex with o-carborane | | Descriptor: | Tetrabrachion, ortho-carborane | | Authors: | Heide, F, McDougall, M, Stetefeld, J. | | Deposit date: | 2021-06-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Boron rich nanotube drug carrier system is suited for boron neutron capture therapy.
Sci Rep, 11, 2021
|
|
6XF4
 
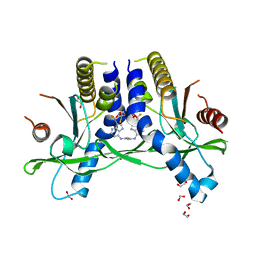 | | Crystal structure of STING REF variant in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), 1,2-ETHANEDIOL, Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6XF3
 
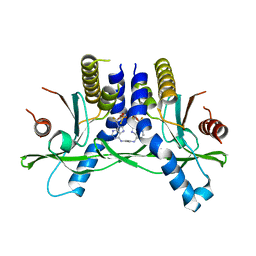 | | Crystal structure of STING in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
8W68
 
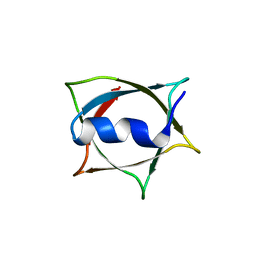 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-08-28 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
5L0T
 
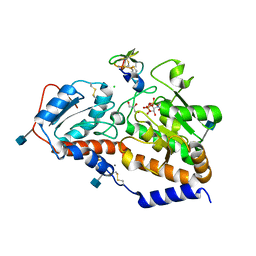 | | human POGLUT1 in complex with EGF(+) and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
6OY4
 
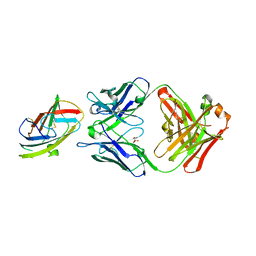 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
6XYR
 
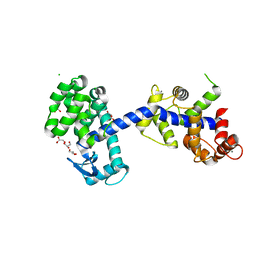 | | Structure of the T4Lnano fusion protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Benoit, R.M, Bierig, T, Collu, C, Engilberge, S, Olieric, V. | | Deposit date: | 2020-01-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Structure, 30, 2022
|
|
5L0U
 
 | | human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EGF(+), ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5GSF
 
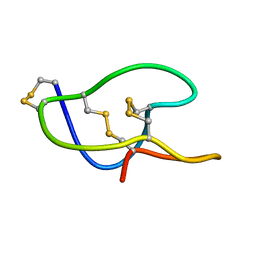 | | Structure of roseltide rT1 | | Descriptor: | roseltide rT1 | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of Roseltide, a Knottin-type Neutrophil Elastase Inhibitor Derived from Hibiscus sabdariffa.
Sci Rep, 6, 2016
|
|
5L0V
 
 | | human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-beta-D-glucopyranose, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5L0S
 
 | | human POGLUT1 in complex with Factor VII EGF1 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
6BVB
 
 | | Crystal structure of HIF-2alpha-pVHL-elongin B-elongin C | | Descriptor: | Elongin-B, Elongin-C, Hypoxia-Inducible Factor 2 alpha, ... | | Authors: | Tarade, D, Ohh, M, Lee, J.E. | | Deposit date: | 2017-12-12 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | HIF-2 alpha-pVHL complex reveals broad genotype-phenotype correlations in HIF-2 alpha-driven disease.
Nat Commun, 9, 2018
|
|
2ZJG
 
 | | Crystal structural of mouse kynurenine aminotransferase III | | Descriptor: | GLYCEROL, Kynurenine-oxoglutarate transaminase 3 | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-03-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of mouse kynurenine aminotransferase III
To be Published
|
|
8TN3
 
 | |
8TN2
 
 | |
