3W5U
 
 | | Cross-linked complex between Ferredoxin and Ferredoxin-NADP+ reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin, ... | | Authors: | Kimata-Ariga, Y, Kubota-Kawai, H, Muraki, N, Hase, T, Kurisu, G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Concentration-dependent oligomerization of cross-linked complexes between ferredoxin and ferredoxin-NADP(+) reductase
Biochem.Biophys.Res.Commun., 434, 2013
|
|
3W5V
 
 | | Cross-linked complex between Ferredoxin and Ferredoxin-NADP+ reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin, ... | | Authors: | Kimata-Ariga, Y, Kubota-Kawai, H, Muraki, N, Hase, T, Kurisu, G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Concentration-dependent oligomerization of cross-linked complexes between ferredoxin and ferredoxin-NADP(+) reductase
Biochem.Biophys.Res.Commun., 434, 2013
|
|
2KFZ
 
 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND ZINC ONLY | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-02 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KFN
 
 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND MANGANESE | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-01 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
4JN2
 
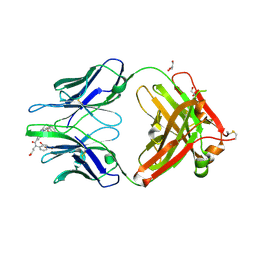 | | An Antidote for Dabigatran | | Descriptor: | GLYCEROL, N-[(2-{[(4-carbamimidoylphenyl)amino]methyl}-1-methyl-1H-benzimidazol-5-yl)carbonyl]-N-pyridin-2-yl-beta-alanine, anti dabigatran Fab | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2013-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A specific antidote for dabigatran: functional and structural characterization.
Blood, 121, 2013
|
|
4JN1
 
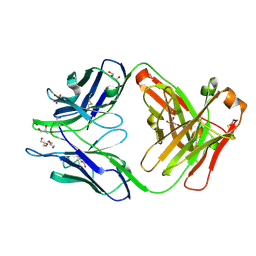 | | An Antidote for Dabigatran | | Descriptor: | GLYCEROL, anti-dabigatran Fab1, heavy chain, ... | | Authors: | Schiele, F, Nar, H. | | Deposit date: | 2013-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A specific antidote for dabigatran: functional and structural characterization.
Blood, 121, 2013
|
|
2KZM
 
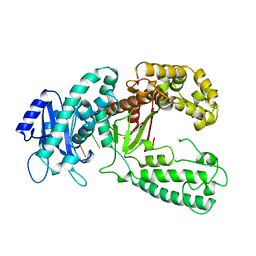 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC AND MANGANESE | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*A*CP*GP*C)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE I), ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-03 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2KZZ
 
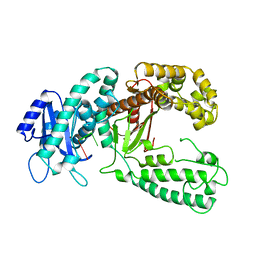 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY | | Descriptor: | DNA (5'-D(*GP*CP*TP*T*AP*CP*G)-3'), PROTEIN (DNA POLYMERASE I), ZINC ION | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-07 | | Release date: | 1999-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
2L5S
 
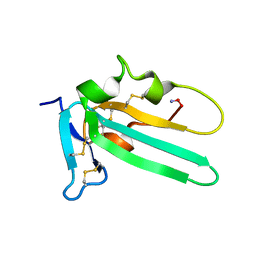 | | Solution structure of the extracellular domain of the TGF-beta type I receptor | | Descriptor: | TGF-beta receptor type-1 | | Authors: | Zuniga, J.E, Ilangovan, U, Pardeep, M, Hinck, C, Huang, T. | | Deposit date: | 2010-11-04 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The TbetaR-I Pre-Helix Extension Is Structurally Ordered in the Unbound Form and Its Flanking Prolines Are Essential for Binding
J.Mol.Biol., 412, 2011
|
|
4HTQ
 
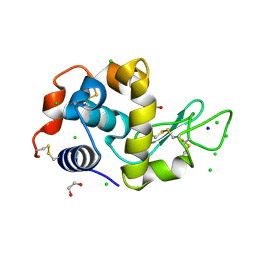 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 6.70 x 10e+11 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.Z, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HTK
 
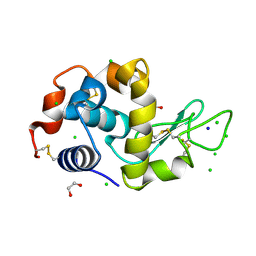 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 2.17 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1CT8
 
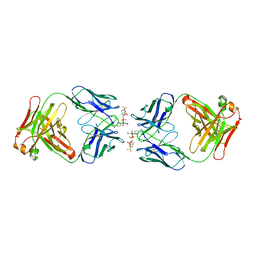 | | CATALYTIC ANTIBODY 7C8 COMPLEX | | Descriptor: | 7C8 FAB FRAGMENT; LONG CHAIN, 7C8 FAB FRAGMENT; SHORT CHAIN, SULFATE ION, ... | | Authors: | Gigant, B, Tsumuraya, T, Fujii, I, Knossow, M. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diverse structural solutions to catalysis in a family of antibodies.
Structure Fold.Des., 7, 1999
|
|
4HTN
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 1.32 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3AQ8
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Q46E mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ9
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Q50E mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
3AQ6
 
 | |
3AQ5
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Fe(II)-O2 form | | Descriptor: | Group 1 truncated hemoglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
2XHE
 
 | | Crystal structure of the Unc18-syntaxin 1 complex from Monosiga brevicollis | | Descriptor: | SYNTAXIN1, UNC18 | | Authors: | Burkhardt, P, Stegmann, C.M, Wahl, M.C, Fasshauer, D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Primordial Neurosecretory Apparatus Identified in the Choanoflagellate Monosiga Brevicollis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AQ7
 
 | | Crystal structure of truncated hemoglobin from Tetrahymena pyriformis, Y25F mutant, Fe(III) form | | Descriptor: | Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Igarashi, J, Kobayashi, K, Matsuoka, A. | | Deposit date: | 2010-10-25 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification.
J.Biol.Inorg.Chem., 16, 2011
|
|
1YEG
 
 | | STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH REACTION PRODUCT | | Descriptor: | ACETATE ION, IGG2A FAB FRAGMENT, PARANITROBENZYL ALCOHOL, ... | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1YEF
 
 | | STRUCTURE OF IGG2A FAB FRAGMENT (D2.3) COMPLEXED WITH SUBSTRATE ANALOGUE | | Descriptor: | IGG2A FAB FRAGMENT, PARA-NITROBENZYL GLUTARYL GLYCINIC ACID, ZINC ION | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1YEH
 
 | | STRUCTURE OF IGG2A FAB FRAGMENT | | Descriptor: | FAB FRAGMENT, ZINC ION | | Authors: | Gigant, B, Knossow, M. | | Deposit date: | 1997-05-29 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | X-ray structures of a hydrolytic antibody and of complexes elucidate catalytic pathway from substrate binding and transition state stabilization through water attack and product release.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
8SKU
 
 | | Structure of human SIgA1 in complex with human CD89 (FcaR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Liu, Q, Stadtmueller, B.M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SIgA structures bound to Streptococcus pyogenes M4 and human CD89 provide insights into host-pathogen interactions.
Nat Commun, 14, 2023
|
|
7PC2
 
 | | HIV-1 Env (BG505 SOSIP.664) in complex with the IgA bNAb 7-269 and the antibody 3BNC117. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC IgG Fab heavy chain, ... | | Authors: | Fernandez, I, Bontems, F, Pehau-Arnaudet, G, Rey, F. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Epitope convergence of broadly HIV-1 neutralizing IgA and IgG antibody lineages in a viremic controller.
J.Exp.Med., 219, 2022
|
|
7TPG
 
 | | Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state | | Descriptor: | Fab Heavy (H) Chain, Fab Light (L) Chain, GERANYL DIPHOSPHATE, ... | | Authors: | Ashraf, K.U, Nygaard, R, Vickery, O.N, Erramilli, S.K, Herrera, C.M, McConville, T.H, Petrou, V.I, Giacometti, S.I, Dufrisne, M.B, Nosol, K, Zinkle, A.P, Graham, C.L.B, Loukeris, M, Kloss, B, Skorupinska-Tudek, K, Swiezewska, E, Roper, D, Clarke, O.B, Uhlemann, A.C, Kossiakoff, A.A, Trent, M.S, Stansfeld, P.J, Mancia, F. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of lipopolysaccharide maturation by the O-antigen ligase.
Nature, 604, 2022
|
|
