1UUV
 
 | | NAPHTHALENE 1,2-DIOXYGENASE WITH NITRIC OXIDE AND INDOLE BOUND IN THE ACTIVE SITE. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2004-01-11 | | Release date: | 2005-02-09 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | No Binding to Naphthalene Dioxygenase.
J.Biol.Inorg.Chem., 10, 2005
|
|
2F1O
 
 | | Crystal Structure of NQO1 with Dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Shaul, Y, Asher, G, Dym, O, Tsvetkov, P, Adler, J, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2005-11-15 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of NAD(P)H quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol.
Biochemistry, 45, 2006
|
|
3PZW
 
 | | Soybean lipoxygenase-1 - re-refinement | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | Authors: | Chruszcz, M, Minor, W. | | Deposit date: | 2010-12-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Determination of protein structures - a series of fortunate events.
Biophys.J., 95, 2008
|
|
3OD4
 
 | | Crystal Structure of Factor Inhibiting HIF-1 Alpha Complexed with Inhibitor | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | King, O.N.F, Bashford-Rogers, R, Maloney, D.J, Jadhav, A, Heightman, T.D, Simeonov, A, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Factor Inhibiting HIF-1 Alpha Complexed with Inhibitor
To be Published
|
|
1DVG
 
 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME; SELELENO-METHIONINE DERIVATIVE, MUTATED AT M51T,M93L,M155L,M191L. | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
2IBZ
 
 | | Yeast Cytochrome BC1 Complex with Stigmatellin | | Descriptor: | 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, Cytochrome b, Cytochrome c1, ... | | Authors: | Hunte, C. | | Deposit date: | 2006-09-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Comparison of Stigmatellin Conformations, Free and Bound to the Photosynthetic Reaction Center and the Cytochrome bc(1) Complex.
J.Mol.Biol., 368, 2007
|
|
1W7C
 
 | | PPLO at 1.23 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Duff, A.P, Cohen, A.E, Ellis, P.J, Guss, J.M. | | Deposit date: | 2004-09-01 | | Release date: | 2006-08-31 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The 1.23 A Structure of Pichia Pastoris Lysyl Oxidase Reveals a Lysine-Lysine Cross-Link
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1WE1
 
 | | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC6803 in complex with heme | | Descriptor: | CHLORIDE ION, Heme oxygenase 1, ISOPROPYL ALCOHOL, ... | | Authors: | Sugishima, M, Migita, C.T, Zhang, X, Yoshida, T, Fukuyama, K. | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme
Eur.J.Biochem., 271, 2004
|
|
1PU4
 
 | | Crystal structure of human vascular adhesion protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salminen, T.A, Airenne, T.T. | | Deposit date: | 2003-06-24 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human vascular adhesion protein-1: unique structural features with functional implications.
Protein Sci., 14, 2005
|
|
1DVE
 
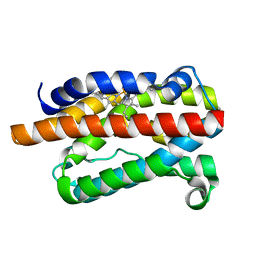 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
1US1
 
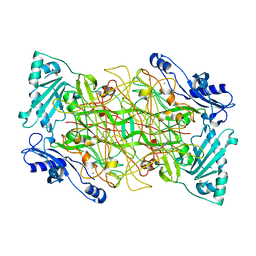 | | Crystal structure of human vascular adhesion protein-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Airenne, T.T, Nymalm, Y, Kidron, H, Soderholm, A, Johnson, M.S, Salminen, T.A. | | Deposit date: | 2003-11-17 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Human Vascular Adhesion Protein-1: Unique Structural Features with Functional Implications.
Protein Sci., 14, 2005
|
|
1SQP
 
 | | Crystal Structure Analysis of Bovine Bc1 with Myxothiazol | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
2DY5
 
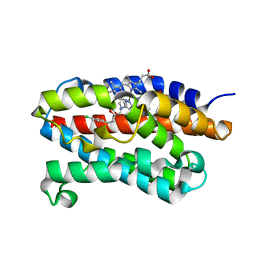 | | Crystal structure of rat heme oxygenase-1 in complex with heme and 2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-1,3-dioxolane | | Descriptor: | 1-({2-[2-(4-CHLOROPHENYL)ETHYL]-1,3-DIOXOLAN-2-YL}METHYL)-1H-IMIDAZOLE, CHLORIDE ION, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Takahashi, H, Fukuyama, K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystallographic and biochemical characterization of the inhibitory action of an imidazole-dioxolane compound on heme oxygenase
Biochemistry, 46, 2007
|
|
2ILM
 
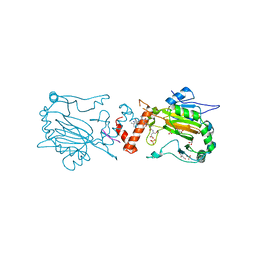 | |
6OD6
 
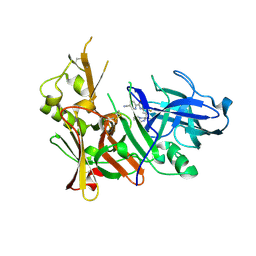 | | Structure of BACE-1 in complex with Ligand 13 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(3R)-1-amino-3-methyl-3,4-dihydropyrrolo[1,2-a]pyrazin-3-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Authors: | Shaffer, P.L. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluation of a Series of beta-Secretase 1 Inhibitors Containing Novel Heteroaryl-Fused-Piperazine Amidine Warheads.
Acs Med.Chem.Lett., 10, 2019
|
|
6OZ2
 
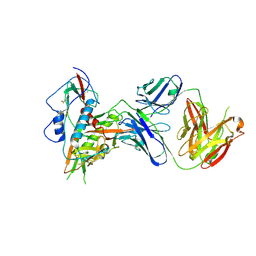 | |
6OQY
 
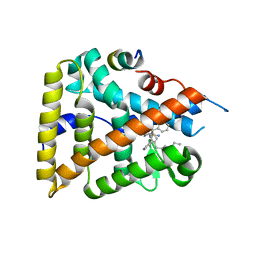 | | Human LRH-1 bound to the agonist 6N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
6OEJ
 
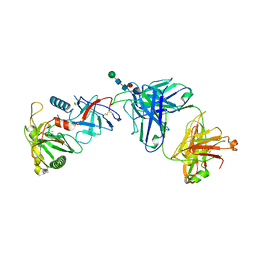 | |
5VN1
 
 | | horse liver alcohol dehydrogenae complexed with NADH (R,S)-N-1-methylhexylformamide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (R)-N-(1-METHYL-HEXYL)-FORMAMIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Plapp, B.V, Ramaswamy, S, Ferraro, D.J, Baskar Raj, S. | | Deposit date: | 2017-04-28 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Horse Liver Alcohol Dehydrogenase: Zinc Coordination and Catalysis.
Biochemistry, 56, 2017
|
|
3K96
 
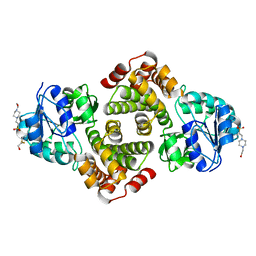 | | 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, Glycerol-3-phosphate dehydrogenase [NAD(P)+] | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Glycerol-3-phosphate Dehydrogenase (gpsA) from Coxiella burnetii.
TO BE PUBLISHED
|
|
3JTW
 
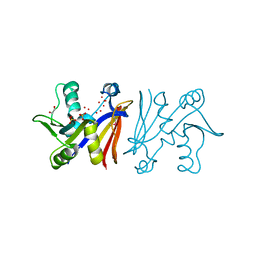 | |
3CH6
 
 | |
8ECU
 
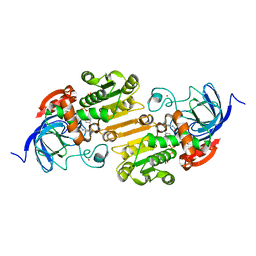 | |
8ECS
 
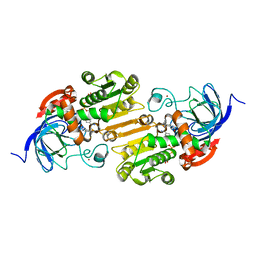 | |
8ECT
 
 | |
