8P8G
 
 | | Nitrogenase MoFe protein from A. vinelandii beta double mutant D353G/D357G | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Maslac, N, Wagner, T. | | Deposit date: | 2023-06-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Mononuclear Metal-Binding Site of Mo-Nitrogenase Is Not Required for Activity.
Jacs Au, 3, 2023
|
|
3L58
 
 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
8OID
 
 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the N111S mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4O03
 
 | | Crystal structure of Ca2+ bound prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-13 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3L5E
 
 | | Structure of BACE Bound to SCH736062 | | Descriptor: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
4MDS
 
 | | Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and non-covalent nanomolar inhibitors with an induced-fit binding | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[4-(acetylamino)phenyl]-2-(1H-benzotriazol-1-yl)-N-[(1R)-2-[(2-methylbutan-2-yl)amino]-1-(1-methyl-1H-pyrrol-2-yl)-2-oxoethyl]acetamide | | Authors: | Mesecar, A.D, Grum-Tokars, V. | | Deposit date: | 2013-08-23 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: Identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M9X
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.344 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
3LC3
 
 | | Benzothiophene Inhibitors of Factor IXa | | Descriptor: | 1-[5-(3,4-dimethoxyphenyl)-1-benzothiophen-2-yl]methanediamine, CALCIUM ION, Coagulation factor IX | | Authors: | Wang, S, Beck, R. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Studies of Benzothiophene Template as Potent Factor IXa (FIXa) Inhibitors in Thrombosis.
J.Med.Chem., 53, 2010
|
|
4NZQ
 
 | | Crystal structure of Ca2+-free prothrombin deletion mutant residues 146-167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Shropshire, D.B, Pelc, L.A, Di Cera, E. | | Deposit date: | 2013-12-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | The linker connecting the two kringles plays a key role in prothrombin activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3IXJ
 
 | | Crystal structure of beta-secretase 1 in complex with selective beta-secretase 1 inhibitor | | Descriptor: | Beta-secretase 1, N-[4-(1-BENZYLCARBAMOYL-2-METHYL-PROPYLCARBAMOYL)-1-(3,5-DIFLUORO-PHENOXYMETHYL)-2-HYDROXY-4-METHOXY-BUTYL]-5-(METHANESULFONYL-METHYL-AMINO)-N'-(1-PHENYLETHYL)-ISOPHTHALAMIDE, SULFATE ION | | Authors: | Borkakoti, N, Lindberg, J, Nystrom, S. | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Potent and Selective BACE-1 Inhibitors.
J.Med.Chem., 53, 2010
|
|
3HY7
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with Marimastat | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3LIW
 
 | | Factor XA in complex with (R)-2-(1-ADAMANTYLCARBAMOYLAMINO)-3-(3-CARBAMIDOYL-PHENYL)-N-PHENETHYL-PROPIONIC ACID AMIDE | | Descriptor: | (R)-2-(3-ADAMANTAN-1-YL-UREIDO)-3-(3-CARBAMIMIDOYL-PHENYL)-N-PHENETHYL-PROPIONAMIDE, Activated factor Xa heavy chain, CALCIUM ION, ... | | Authors: | Mueller, M.M, Sperl, S, Sturzebecher, J, Bode, W, Moroder, L. | | Deposit date: | 2010-01-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | (R)-3-Amidinophenylalanine-Derived Inhibitors of Factor Xa with a Novel Active-Site Binding Mode
Biol.Chem., 383, 2003
|
|
3LNK
 
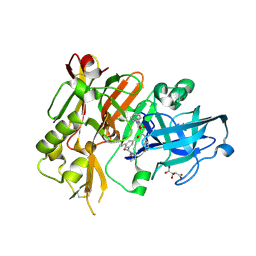 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L5D
 
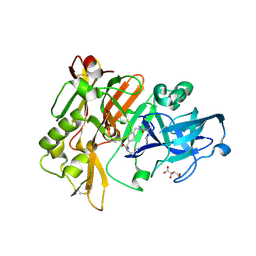 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5B
 
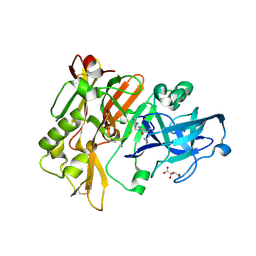 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3KYO
 
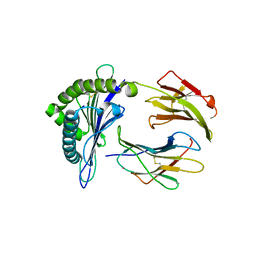 | | Crystal structure of HLA-G presenting KLPAQFYIL peptide | | Descriptor: | Beta-2-microglobulin, COBALT (II) ION, KLPAQFYIL peptide, ... | | Authors: | Walpole, N.G, Rossjohn, J, Clements, C.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure and stability of the monomorphic HLA-G are influenced by the nature of the bound peptide
J.Mol.Biol., 397, 2010
|
|
3LC5
 
 | | Selective Benzothiophine Inhibitors of Factor IXa | | Descriptor: | 1-{4-[(R)-phenyl(3-phenyl-1,2,4-oxadiazol-5-yl)methoxy]-1-benzothiophen-2-yl}methanediamine, CALCIUM ION, Coagulation factor IX | | Authors: | Wang, S, Beck, R. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure Based Drug Design: Development of Potent and Selective Factor IXa (FIXa) Inhibitors.
J.Med.Chem., 53, 2010
|
|
4NIY
 
 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) complexed to YRH-ecotin (M84Y/M85R/A86H ecotin) | | Descriptor: | CALCIUM ION, Cationic trypsin, Ecotin, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5F
 
 | | Structure of BACE Bound to SCH736201 | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3LU9
 
 | | Crystal structure of human thrombin mutant S195A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Proteinase-activated receptor 1, ... | | Authors: | Gandhi, P.S, Chen, Z, Di Cera, E. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1.
J.Biol.Chem., 285, 2010
|
|
3LMS
 
 | | Structure of human activated thrombin-activatable fibrinolysis inhibitor, TAFIa, in complex with tick-derived funnelin inhibitor, TCI. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Carboxypeptidase B2, ... | | Authors: | Sanglas, L, Arolas, J.L, Valnickova, Z, Aviles, F.X, Enghild, J.J, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-01 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the molecular inactivation mechanism of human activated thrombin-activatable fibrinolysis inhibitor
J.Thromb.Haemost., 8, 2010
|
|
5HSP
 
 | | MamM CTD M250L | | Descriptor: | Magnetosome protein MamM, SULFATE ION | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2016-01-26 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disease-Homologous Mutation in the Cation Diffusion Facilitator Protein MamM Causes Single-Domain Structural Loss and Signifies Its Importance.
Sci Rep, 6, 2016
|
|
2VVU
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[(3R)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)pyrrolidin-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-11 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
4TX4
 
 | | Crystal Structure of a Single-Domain Cysteine Protease Inhibitor from Cowpea (Vigna unguiculata) | | Descriptor: | Cysteine proteinase inhibitor, SULFATE ION | | Authors: | Pereira, H.M, Valadares, N, Monteiro-Junior, J.E, Carvalho, C.P.S, Grangeiro, T.B. | | Deposit date: | 2014-07-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression in Escherichia coli of cysteine protease inhibitors from cowpea (Vigna unguiculata): The crystal structure of a single-domain cystatin gives insights on its thermal and pH stability.
Int. J. Biol. Macromol., 102, 2017
|
|
