1KB9
 
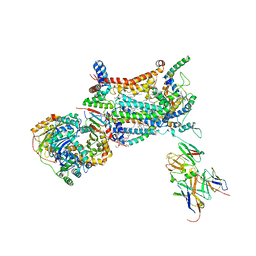 | | YEAST CYTOCHROME BC1 COMPLEX | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Lange, C, Nett, J.H, Trumpower, B.L, Hunte, C. | | Deposit date: | 2001-11-05 | | Release date: | 2002-09-18 | | Last modified: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SPECIFIC ROLES OF PROTEIN-PHOSPHOLIPID INTERACTIONS IN THE YEAST CYTOCHROME BC1 COMPLEX STRUCTURE
EMBO J., 20, 2001
|
|
1OIU
 
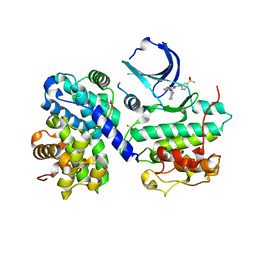 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 3-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
2W06
 
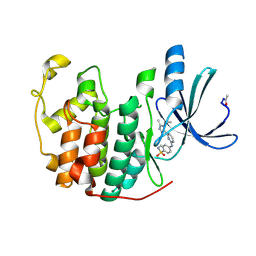 | | Structure of CDK2 in complex with an imidazolyl pyrimidine, compound 5c | | Descriptor: | 4-{[4-(1-CYCLOPROPYL-2-METHYL-1H-IMIDAZOL-5-YL)PYRIMIDIN-2-YL]AMINO}-N-METHYLBENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Anderson, M, Andrews, D.M, Barker, A.J, Brassington, C.A, Byth, K.F, Culshaw, J.D, Finlay, M.R.V, Fisher, E, Mcmiken, H.H.J, Green, C.P, Heaton, D.W, Nash, I.A, Newcombe, N.J, Oakes, S.E, Roberts, A, Stanway, J.J, Thomas, A.P, Tucker, J.A, Weir, H.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Imidazoles: Sar and Development of a Potent Class of Cyclin-Dependent Kinase Inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5A3Q
 
 | | Crystal structure of the (SR) Calcium ATPase E2-vanadate complex bound to thapsigargin and TNP-AMPPCP | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
5A3R
 
 | | Crystal structure of the (SR) Calcium ATPase E2.BeF3- complex bound to TNP-AMPPCP | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SARCOPLASMIC/ENDOPLASMIC RETICULUM CALCIUM ATPASE 1, ... | | Authors: | Clausen, J.D, Bublitz, M, Arnou, B, Olesen, C, Andersen, J.P, Moller, J.V, Nissen, P. | | Deposit date: | 2015-06-02 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal Structure of the Vanadate-Inhibited Ca(2+)-ATPase.
Structure, 24, 2016
|
|
7TML
 
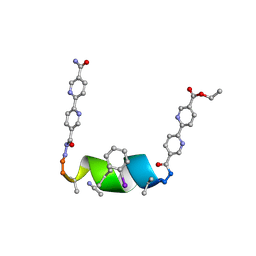 | | Porous framework formed by assembly of a bipyridyl-conjugated helical peptide | | Descriptor: | 5'-(hydrazinecarbonyl)[2,2'-bipyridine]-5-carboxamide, ACETONITRILE, ethyl 5'-formyl[2,2'-bipyridine]-5-carboxylate, ... | | Authors: | Nguyen, A.I. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Assembly of pi-Stacking Helical Peptides into a Porous and Multivariable Proteomimetic Framework.
J.Am.Chem.Soc., 144, 2022
|
|
3NP7
 
 | | Glycogen phosphorylase complexed with 2,5-dihydroxy-3-(beta-D-glucopyranosyl)-chlorobenzene and 2,5-dihydroxy-4-(beta-D-glucopyranosyl)-chlorobenzene | | Descriptor: | (1S)-1,5-anhydro-1-(3-chloro-2,5-dihydroxyphenyl)-D-glucitol, (1S)-1,5-anhydro-1-(4-chloro-2,5-dihydroxyphenyl)-D-glucitol, Glycogen phosphorylase, ... | | Authors: | Alexacou, K.-M. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Halogen-substituted (C-beta-D-glucopyranosyl)-hydroquinone regioisomers: synthesis, enzymatic evaluation and their binding to glycogen phosphorylase.
Bioorg.Med.Chem., 19, 2011
|
|
2WI4
 
 | | Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone | | Descriptor: | 4-(2,4-dichlorophenyl)-5-phenyldiazenyl-pyrimidin-2-amine, HEAT SHOCK PROTEIN, HSP 90-ALPHA | | Authors: | Brough, P.A, Barril, X, Borgognoni, J, Chene, P, Davies, N.G.M, Davis, B, Drysdale, M.J, Dymock, B, Eccles, S.A, Garcia-Echeverria, C, Fromont, C, Hayes, A, Hubbard, R.E, Jordan, A.M, Rugaard-Jensen, M, Massey, A, Merret, A, Padfield, A, Parsons, R, Radimerski, T, Raynaud, F.I, Robertson, A, Roughley, S.D, Schoepfer, J, Simmonite, H, Surgenor, A, Valenti, M, Walls, S, Webb, P, Wood, M, Workman, P, Wright, L.M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combining Hit Identification Strategies: Fragment- Based and in Silico Approaches to Orally Active 2-Aminothieno[2,3-D]Pyrimidine Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 52, 2009
|
|
2XAB
 
 | | Structure of HSP90 with an inhibitor bound | | Descriptor: | 4-(1,3-DIHYDRO-2H-ISOINDOL-2-YLCARBONYL)-6-(1-METHYLETHYL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, m, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, O'Brian, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
1SCM
 
 | |
2LP8
 
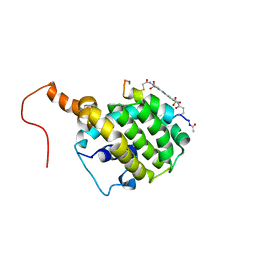 | | SOLUTION STRUCTURE OF AN APOPTOSIS ACTIVATING PHOTOSWITCHABLE BAK PEPTIDE BOUND to BCL-XL | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1 | | Authors: | Wysoczanski, P, Mart, R.J, Loveridge, J.E, Williams, C, Whittaker, S.B.-M, Crump, M.P, Allemann, R.K. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-18 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
J.Am.Chem.Soc., 134, 2012
|
|
5DAC
 
 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with DNA | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
5DBY
 
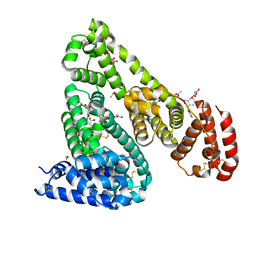 | | Crystal Structure of Equine Serum Albumin in Complex with Diclofenac and Naproxen Obtained in Displacement Experiment | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ... | | Authors: | Sekula, B, Bujacz, A, Bujacz, G. | | Deposit date: | 2015-08-22 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insights into the Competitive Binding of Diclofenac and Naproxen by Equine Serum Albumin.
J.Med.Chem., 59, 2016
|
|
5DFD
 
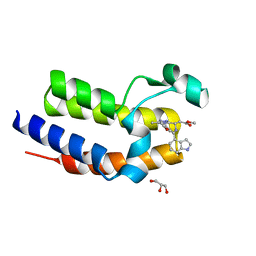 | | Crystal structure of BRD2(BD2) W370F mutant with ligand 28 bound | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, methyl [(4S)-6-(1H-indol-4-yl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]acetate | | Authors: | Tallant, C, Baud, M, Lin-Shiao, E, Chirgadze, D.Y, Ciulli, A. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Synthetic Routes to Triazolo-benzodiazepine Analogues: Expanding the Scope of the Bump-and-Hole Approach for Selective Bromo and Extra-Terminal (BET) Bromodomain Inhibition.
J.Med.Chem., 59, 2016
|
|
8F1S
 
 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in A-orientation | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
5CW5
 
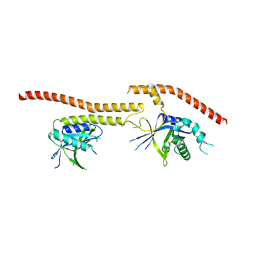 | | Structure of CfBRCC36-CfKIAA0157 complex (QSQ mutant) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.736 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
8F0F
 
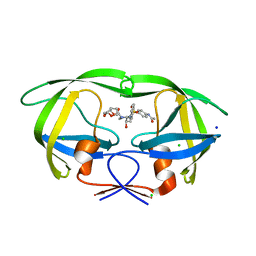 | | HIV-1 wild type protease with GRL-110-19A, a chloroacetamide derivative based on Darunavir as P2' group | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[4-(2-chloroacetamido)benzene-1-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2' amide-derivatives: Synthesis, biological evaluation and structural studies.
Bioorg.Med.Chem.Lett., 83, 2023
|
|
5CW3
 
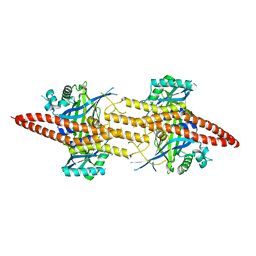 | | Structure of CfBRCC36-CfKIAA0157 complex (Zn Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
8F1V
 
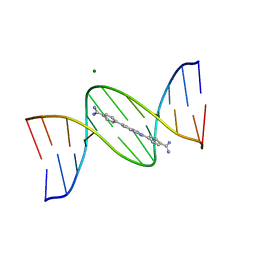 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in B-orientation | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
5D6C
 
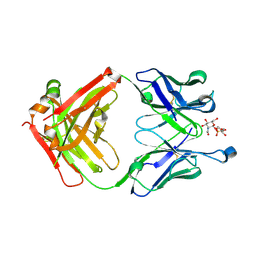 | | Structure of 4497 Fab bound to synthetic wall teichoic acid fragment | | Descriptor: | 4-O-[2-acetamido-2-deoxy-beta-D-glucopyranosyl]-5-O-phosphono-D-ribitol, 4497 antibody IgG1 (VH and CH1), 4497 antibody IgK (VL and CL), ... | | Authors: | Lupardus, P.J, Fong, R. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel antibody-antibiotic conjugate eliminates intracellular S. aureus.
Nature, 527, 2015
|
|
8EQZ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-hexylbenzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2022-10-11 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8F0S
 
 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-9296 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0P
 
 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the hybrid inhibitor GNE-1305 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
8F0R
 
 | | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the arylsulfonamide inhibitor GNE-3565 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-4-({(1S,2S,4S)-2-(dimethylamino)-4-[3-(trifluoromethyl)phenyl]cyclohexyl}amino)-2-fluoro-N-(pyrimidin-4-yl)benzene-1-sulfonamide, ... | | Authors: | Kschonsak, M, Jao, C.C, Arthur, C.P, Rohou, A.L, Bergeron, P, Ortwine, D, McKerall, S.J, Hackos, D.H, Deng, L, Chen, J, Sutherlin, D, Dragovich, P.S, Volgraf, M, Wright, M.R, Payandeh, J, Ciferri, C, Tellis, J.C. | | Deposit date: | 2022-11-03 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals an unprecedented binding site for Na V 1.7 inhibitors enabling rational design of potent hybrid inhibitors.
Elife, 12, 2023
|
|
5DFC
 
 | | Crystal structure of BRD2(BD2) W370F mutant with ligand I-BET 762 bound | | Descriptor: | 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide, Bromodomain-containing protein 2, GLYCEROL, ... | | Authors: | Tallant, C, Baud, M, Lin-Shiao, E, Chirgadze, D.Y, Ciulli, A. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New Synthetic Routes to Triazolo-benzodiazepine Analogues: Expanding the Scope of the Bump-and-Hole Approach for Selective Bromo and Extra-Terminal (BET) Bromodomain Inhibition.
J.Med.Chem., 59, 2016
|
|
