7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
8BL4
 
 | | Cryo-EM structure of a contractile injection system in Streptomyces coelicolor, the sheath-tube module in extended state. | | Descriptor: | Phage tail protein, Phage tail sheath family protein | | Authors: | Casu, B, Sallmen, J.W, Schlimpert, S, Pilhofer, M. | | Deposit date: | 2022-11-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cytoplasmic contractile injection systems mediate cell death in Streptomyces.
Nat Microbiol, 8, 2023
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8BSJ
 
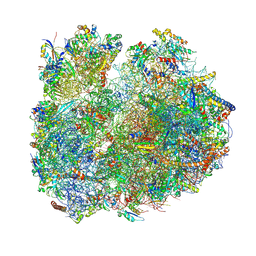 | | Giardia Ribosome in PRE-T Classical State (C) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-25 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (6.49 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
6VYY
 
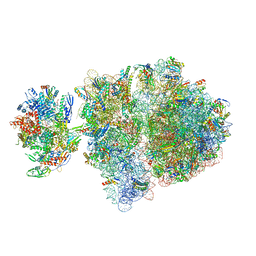 | | Escherichia coli transcription-translation complex C5 (TTC-C5) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
7SUK
 
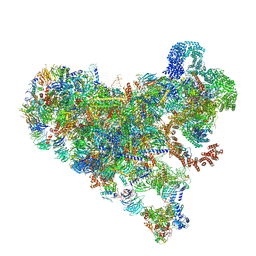 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | Descriptor: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-11-17 | | Release date: | 2022-07-06 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
6W0D
 
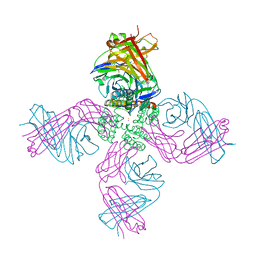 | | Open-gate KcsA soaked in 5 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.639 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6VYS
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing a 21 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6W0J
 
 | | Closed-gate KcsA incubated in BaCl2/NaCl | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
8BHJ
 
 | |
6W1D
 
 | | Structure of human mitochondrial complex Nfs1-ISCU2 (WT)-ISD11 with E.coli ACP1 at 1.8 A resolution (NIAU)2 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
6W1O
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8BHN
 
 | |
8BIM
 
 | |
8BGH
 
 | |
2G9T
 
 | | Crystal structure of the SARS coronavirus nsp10 at 2.1A | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Yang, H, Sun, F, Rao, Z. | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
8BF7
 
 | |
8BUM
 
 | | Structure of DDB1 bound to DS15-engaged CDK12-cyclin K | | Descriptor: | (2R)-2-[[6-(5-naphthalen-1-ylpentylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
6WP0
 
 | |
8BIL
 
 | |
6WQ0
 
 | | Cryo-EM of the S. solfataricus rod-shaped virus, SSRV1 | | Descriptor: | DNA (301-MER), Structural protein | | Authors: | Wang, F, Baquero, D.P, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SYX
 
 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-13 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6WX6
 
 | |
8BU7
 
 | | Structure of DDB1 bound to 21195-engaged CDK12-cyclin K | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(chloranyl)phenyl]-6-[[4-(2-hydroxyethyloxy)phenyl]methyl]-3-propan-2-yl-5H-pyrazolo[3,4-d]pyrimidin-4-one, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.245 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUN
 
 | | Structure of DDB1 bound to DS16-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-[(4-phenylphenyl)methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
