1L0E
 
 | |
1PTX
 
 | |
1T87
 
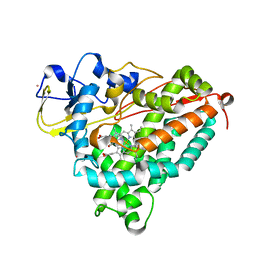 | | Crystal Structure of the Ferrous CO-bound Cytochrome P450cam (C334A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CARBON MONOXIDE, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1T88
 
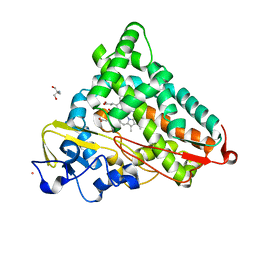 | | Crystal Structure of the Ferrous Cytochrome P450cam (C334A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Cytochrome P450-cam, ... | | Authors: | Nagano, S, Tosha, T, Ishimori, K, Morishima, I, Poulos, T.L. | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding.
J.Biol.Chem., 279, 2004
|
|
1PI5
 
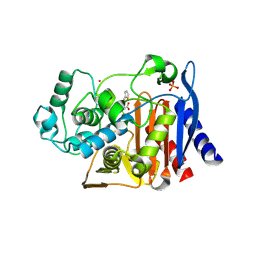 | | Structure of N289A mutant of AmpC in complex with SM2, carboxyphenylglycylboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Roth, T.A, Minasov, G, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2003-05-29 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Thermodynamic cycle analysis and inhibitor design against beta-lactamase.
Biochemistry, 42, 2003
|
|
7SZO
 
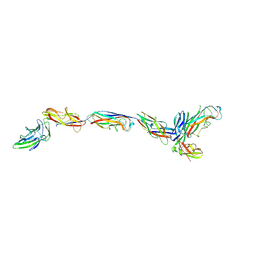 | | Structure of a bacterial fimbrial tip containing FocH | | Descriptor: | Chaperone protein FimC, FimF protein, FimG, ... | | Authors: | Stenkamp, R.E, Le Trong, I, Aprikian, P, Sokurenko, E.V. | | Deposit date: | 2021-11-29 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant FimH Adhesin Demonstrates How the Allosteric Catch Bond Mechanism Can Support Fast and Strong Bacterial Attachment in the Absence of Shear.
J.Mol.Biol., 434, 2022
|
|
8DPA
 
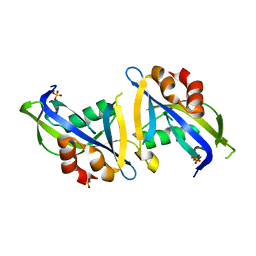 | | Crystal structure of the homodimeric AvrM14-B Nudix hydrolase effector from Melampsora lini | | Descriptor: | AvrM14-B, SULFATE ION | | Authors: | McCombe, C.L, Outram, M.A, Ericsson, D.J, Williams, S.J. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A rust-fungus Nudix hydrolase effector decaps mRNA in vitro and interferes with plant immune pathways.
New Phytol., 239, 2023
|
|
8DP9
 
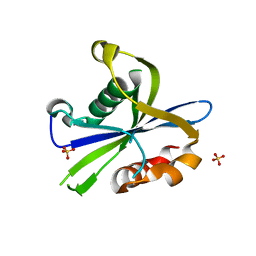 | | Crystal structure of the monomeric AvrM14-B Nudix hydrolase effector from Melampsora lini | | Descriptor: | AvrM14-B, SULFATE ION | | Authors: | McCombe, C.L, Outram, M.A, Ericsson, D.J, Williams, S.J. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A rust-fungus Nudix hydrolase effector decaps mRNA in vitro and interferes with plant immune pathways.
New Phytol., 239, 2023
|
|
8DP8
 
 | |
3FWG
 
 | | Ferric camphor bound Cytochrome P450cam, Arg365Leu, Glu366Gln, monoclinic crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Schlichting, I, Von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7LCU
 
 | | X-ray structure of Furin bound to BOS-318, a small molecule inhibitor | | Descriptor: | (1-{[2-(3,5-dichlorophenyl)-6-{[2-(4-methylpiperazin-1-yl)pyrimidin-5-yl]oxy}pyridin-4-yl]methyl}piperidin-4-yl)acetic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Campobasso, N, Reid, R. | | Deposit date: | 2021-01-11 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A highly selective, cell-permeable furin inhibitor BOS-318 rescues key features of cystic fibrosis airway disease.
Cell Chem Biol, 29, 2022
|
|
8E37
 
 | | Structure of Campylobacter concisus wild-type SeMet PglC | | Descriptor: | N,N'-diacetylbacilliosaminyl-1-phosphate transferase | | Authors: | Vuksanovic, N, Ray, L.C, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Synergistic computational and experimental studies of a phosphoglycosyl transferase membrane/ligand ensemble.
J.Biol.Chem., 299, 2023
|
|
7ADQ
 
 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
1PI4
 
 | | Structure of N289A mutant of AmpC in complex with SM3, a phenylglyclboronic acid bearing the cephalothin R1 side chain | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Roth, T.A, Minasov, G, Focia, P.J, Shoichet, B.K. | | Deposit date: | 2003-05-29 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Thermodynamic cycle analysis and inhibitor design against beta-lactamase.
Biochemistry, 42, 2003
|
|
2L9G
 
 | |
7KEL
 
 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59K mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Sanders, D.A.R. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reversing the roles of a crucial hydrogen-bonding pair: a lysine-insensitive mutant of Campylobacter jejuni dihydrodipicolinate synthase, H59K, binds histidine in its allosteric site
To Be Published
|
|
7KG9
 
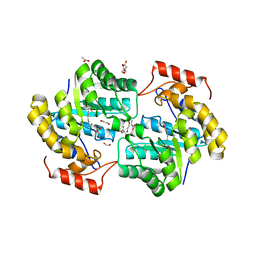 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site and L-lysine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
7KG2
 
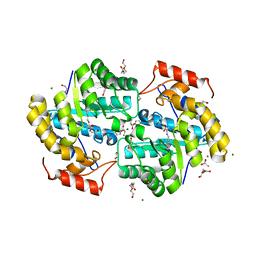 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H59K mutant with pyruvate bound in the active site and L-histidine bound at the allosteric site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Reversing the roles of a crucial hydrogen-bonding pair: a lysine-insensitive mutant of Campylobacter jejuni dihydrodipicolinate synthase, H59K, binds histidine in its allosteric site
To be Published
|
|
7KG5
 
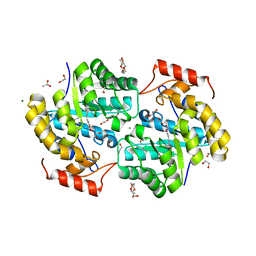 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ACETATE ION, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
7KH4
 
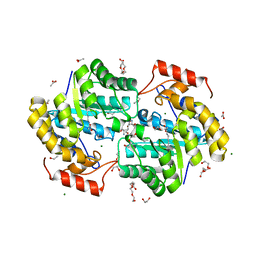 | | Dihydrodipicolinate synthase (DHDPS) from C.jejuni, H56W mutant with pyruvate bound in the active site and R,R-bislysine bound at the allosteric site | | Descriptor: | (2R,5R)-2,5-diamino-2,5-bis(4-aminobutyl)hexanedioic acid, 1,2-ETHANEDIOL, 4-hydroxy-tetrahydrodipicolinate synthase, ... | | Authors: | Saran, S, Majdi Yazdi, M, Sanders, D.A.R. | | Deposit date: | 2020-10-19 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
To Be Published
|
|
1O07
 
 | | Crystal Structure of the complex between Q120L/Y150E mutant of AmpC and a beta-lactam inhibitor (MXG) | | Descriptor: | 2-(1-{2-[4-(2-ACETYLAMINO-PROPIONYLAMINO)-4-CARBOXY-BUTYRYLAMINO]-6-AMINO-HEXANOYLAMINO}-2-OXO-ETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, POTASSIUM ION | | Authors: | Meroueh, S.O, Minasov, G, Lee, W, Shoichet, B.K, Mobashery, S. | | Deposit date: | 2003-02-20 | | Release date: | 2003-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Aspects for Evolution of beta-Lactamases from Penicillin-Binding Proteins
J.Am.Chem.Soc., 125, 2003
|
|
2QVT
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-D | | Descriptor: | AvrL567-D | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Lawrence, G, Schirra, H.J, Anderson, P.A, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity.
Plant Cell, 19, 2007
|
|
6T6W
 
 | |
6TC1
 
 | |
6TGL
 
 | |
