1OVA
 
 | |
3OC7
 
 | |
5T41
 
 | |
5T16
 
 | |
3RR6
 
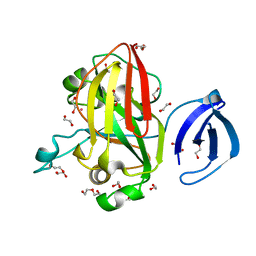 | |
3R59
 
 | | Human Cyclophilin D Complexed with a Fragment | | Descriptor: | 1-(3-aminophenyl)ethanone, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
2TEC
 
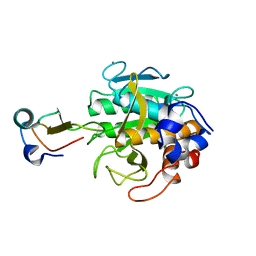 | | MOLECULAR DYNAMICS REFINEMENT OF A THERMITASE-EGLIN-C COMPLEX AT 1.98 ANGSTROMS RESOLUTION AND COMPARISON OF TWO CRYSTAL FORMS THAT DIFFER IN CALCIUM CONTENT | | Descriptor: | CALCIUM ION, EGLIN C, THERMITASE | | Authors: | Gros, P, Betzel, C, Dauter, Z, Wilson, K.S, Hol, W.G.J. | | Deposit date: | 1990-10-26 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular dynamics refinement of a thermitase-eglin-c complex at 1.98 A resolution and comparison of two crystal forms that differ in calcium content.
J.Mol.Biol., 210, 1989
|
|
2BEM
 
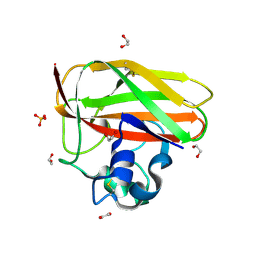 | | Crystal structure of the Serratia marcescens chitin-binding protein CBP21 | | Descriptor: | 1,2-ETHANEDIOL, CBP21, SODIUM ION, ... | | Authors: | Vaaje-Kolstad, G, Houston, D.R, Eijsink, V.G.H, van Aalten, D.M.F. | | Deposit date: | 2004-11-26 | | Release date: | 2004-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure and Binding Properties of the Serratia Marcescens Chitin-Binding Protein Cbp21
J.Biol.Chem., 280, 2005
|
|
2C4X
 
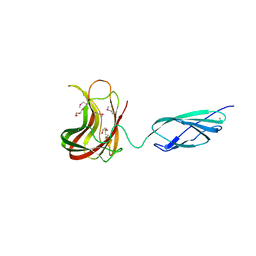 | | Structural basis for the promiscuous specificity of the carbohydrate- binding modules from the beta-sandwich super family | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOGLUCANASE | | Authors: | Najmudin, S, Guerreiro, C.I.P.D, Carvalho, A.L, Bolam, D.N, Prates, J.A.M, Correia, M.A.S, Alves, V.D, Ferreira, L.M.A, Romao, M.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2005-10-25 | | Release date: | 2005-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Xyloglucan is Recognized by Carbohydrate-Binding Modules that Interact with Beta-Glucan Chains.
J.Biol.Chem., 281, 2006
|
|
2ST1
 
 | |
8CA3
 
 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
7H1U
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z212053854 | | Descriptor: | 1,2,3,4-tetrahydro-1,5-naphthyridine, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
5CEQ
 
 | |
3HLZ
 
 | |
3HM5
 
 | | SANT domain of human DNA methyltransferase 1 associated protein 1 | | Descriptor: | CALCIUM ION, DNA methyltransferase 1-associated protein 1, UNKNOWN ATOM OR ION | | Authors: | Dombrovski, L, Tempel, W, Amaya, M.F, Tong, Y, Ni, S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Park, H, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SANT domain of human DNA methyltransferase 1 associated protein 1
To be Published
|
|
7H1M
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with POB0075 | | Descriptor: | 1-[1-(pyridin-2-yl)cyclopentyl]methanamine, DIMETHYL SULFOXIDE, Serine protease NS3, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
1YQ7
 
 | | Human farnesyl diphosphate synthase complexed with risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION, ... | | Authors: | Kavanagh, K.L, Guo, K, Von delft, F, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-02-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human farnesyl diphosphate complexed with clinical inhibitor risedronate
To be Published
|
|
2D1A
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the extended-duplex dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
5COR
 
 | |
4J2U
 
 | | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1 | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1
To be Published
|
|
4ESP
 
 | | Crystal Structure of Peanut Allergen Ara h 5 | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Profilin | | Authors: | Wang, Y, Zhang, Y.Z. | | Deposit date: | 2012-04-23 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of Peanut ( Arachis hypogaea ) Allergen Ara h 5.
J.Agric.Food Chem., 61, 2013
|
|
5FL6
 
 | | Three dimensional structure of human carbonic anhydrase IX in complex with 5-(1-(4-Methylphenyl)-1H-1,2,3-triazol-4-yl)thiophene-2- sulfonamide | | Descriptor: | 5-[1-(4-methylphenyl)-1,2,3-triazol-4-yl]thiophene-2-sulfonamide, ACETIC ACID, CARBONIC ANHYDRASE IX, ... | | Authors: | Leitans, J, Tars, K, Zalubovskis, R. | | Deposit date: | 2015-10-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Efficient Expression and Crystallization System of the Cancer Asociated Carbonic Anhydrase Isoform Ix.
J.Med.Chem., 58, 2015
|
|
5MU9
 
 | | MOA-E-64 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Agglutinin, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Family of Papain-Like Fungal Chimerolectins with Distinct Ca(2+)-Dependent Activation Mechanism.
Biochemistry, 56, 2017
|
|
3TVL
 
 | | Complex between the human thiamine triphosphatase and triphosphate | | Descriptor: | 1,2-ETHANEDIOL, TRIPHOSPHATE, Thiamine-triphosphatase | | Authors: | Delvaux, D, Herman, R, Sauvage, E, Wins, P, Bettendorff, L, Charlier, P, Kerff, F. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase.
Biochim.Biophys.Acta, 1830, 2013
|
|
6HVW
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with 43 | | Descriptor: | (2~{S})-~{N}-[(2~{S},3~{R})-1-[(4~{a}~{S},8~{a}~{S})-1,2,3,4,4~{a},5,6,7,8,8~{a}-decahydronaphthalen-2-yl]-4-methyl-3,4-bis(oxidanyl)pentan-2-yl]-3-(4-methoxyphenyl)-2-[[(2~{S})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
