8VK9
 
 | | Structure of UbV.d2.3 in complex with Ube2d2-S22R | | Descriptor: | GLYCEROL, Ubiquitin variant D2.3, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Middleton, A.J. | | Deposit date: | 2024-01-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biophysical characterisation of ubiquitin variants that inhibit the ubiquitin conjugating enzyme Ube2d2.
Febs J., 291, 2024
|
|
8SYC
 
 | | Crystal structure of PDE3B in complex with GSK4394835A | | Descriptor: | MAGNESIUM ION, [3-[(4,7-dimethoxyquinolin-2-yl)carbonylamino]-5-[methyl-(phenylmethyl)carbamoyl]phenyl]-oxidanyl-oxidanylidene-boron, cGMP-inhibited 3',5'-cyclic phosphodiesterase 3B | | Authors: | Concha, N.O, Nolte, R. | | Deposit date: | 2023-05-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and SAR Study of Boronic Acid-Based Selective PDE3B Inhibitors from a Novel DNA-Encoded Library.
J.Med.Chem., 67, 2024
|
|
8T1M
 
 | | Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Agdanowski, M.P, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8IVL
 
 | | FABP7 complexed with Cholesterol | | Descriptor: | CHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
8IVF
 
 | | FABP7 complexed with 25-HC | | Descriptor: | 25-HYDROXYCHOLESTEROL, Fatty acid-binding protein, brain | | Authors: | Wei, P.C, Zhao, K, Yin, L. | | Deposit date: | 2023-03-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fatty acid-binding proteins 3, 7, and 8 bind cholesterol and facilitate its egress from lysosomes.
J.Cell Biol., 223, 2024
|
|
8BA0
 
 | | Drosophila melanogaster complex I in the Twisted state (Dm2) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Agip, A.N.A, Chung, I, Sanchez-Martinez, A, Whitworth, A.J, Hirst, J. | | Deposit date: | 2022-10-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structures of mitochondrial respiratory complex I from Drosophila melanogaster.
Elife, 12, 2023
|
|
8B9Z
 
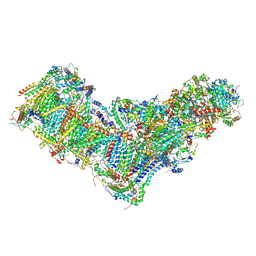 | | Drosophila melanogaster complex I in the Active state (Dm1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Agip, A.A, Chung, I, Sanchez-Martinez, A, Whitworth, A.J, Hirst, J. | | Deposit date: | 2022-10-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures of mitochondrial respiratory complex I from Drosophila melanogaster.
Elife, 12, 2023
|
|
4WBH
 
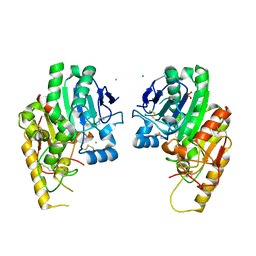 | |
4WDI
 
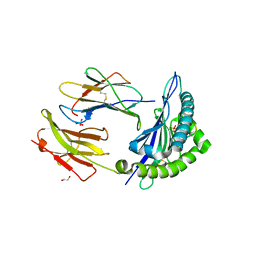 | |
6M14
 
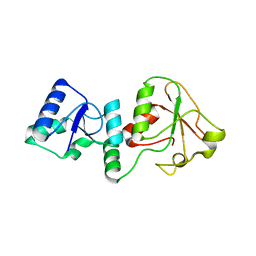 | | Crystal Structure of the BARD1 BRCT Mutant | | Descriptor: | BRCA1-associated RING domain protein 1, SULFATE ION | | Authors: | Chen, T, Huang, W.T. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88001835 Å) | | Cite: | BARD1 is an ATPase activating protein for OLA1.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
8VJT
 
 | | Structure of the poly-UG quadruplex (GUGUGU)4 | | Descriptor: | POTASSIUM ION, RNA (5'-R(*GP*UP*GP*UP*GP*U)-3'), SODIUM ION | | Authors: | Bingman, C.A, Montemayor, E.J, Roschdi, S, Nomura, Y, Butcher, S.E. | | Deposit date: | 2024-01-08 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | Self-assembly and condensation of intermolecular poly(UG) RNA quadruplexes.
Nucleic Acids Res., 52, 2024
|
|
6M0Q
 
 | | Hydroxylamine oxidoreductase from Nitrosomonas europaea | | Descriptor: | Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
6M0P
 
 | | Hydroxylamine oxidoreductase in complex with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, Aerobic hydroxylamine oxidoreductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fujiwara, T, Fujimoto, Z, Nishigaya, Y, Yamazaki, T. | | Deposit date: | 2020-02-22 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Juglone, a plant-derived 1,4-naphthoquinone, binds to hydroxylamine oxidoreductase and inhibits the electron transfer to cytochrome c 554.
Appl.Environ.Microbiol., 89, 2023
|
|
4UUI
 
 | |
8FU5
 
 | | Crystal structure of Fe-CAO1 in complex with piceatannol | | Descriptor: | BENZOIC ACID, Carotenoid oxygenase, FE (II) ION, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2023-01-16 | | Release date: | 2024-08-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Spectroscopy and crystallography define carotenoid oxygenases as a new subclass of mononuclear non-heme Fe II enzymes.
J.Biol.Chem., 2025
|
|
8FU2
 
 | | Crystal structure of T151G CAO1 in complex with piceatannol | | Descriptor: | BENZOIC ACID, Carotenoid oxygenase, FE (II) ION, ... | | Authors: | Kiser, P.D. | | Deposit date: | 2023-01-16 | | Release date: | 2024-08-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spectroscopy and crystallography define carotenoid oxygenases as a new subclass of mononuclear non-heme Fe II enzymes.
J.Biol.Chem., 2025
|
|
4URG
 
 | | Crystal Structure of GGDEF domain from T.maritima (active-like dimer) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DIGUANYLATE CYCLASE | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swaminathan, K, Lescar, J. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
6M2D
 
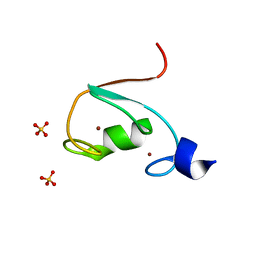 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
8V2U
 
 | |
6M2C
 
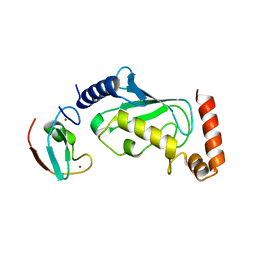 | |
8V2M
 
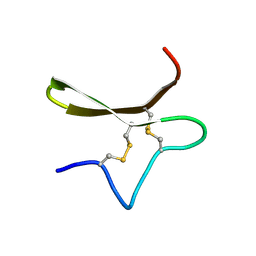 | | Structure of Asterias rubens peptide (KASH2) | | Descriptor: | Kartesh 2 | | Authors: | Takjoo, R, Daly, N.L. | | Deposit date: | 2023-11-23 | | Release date: | 2024-11-27 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of an Asterias rubens peptide indicates the presence of a disulfide-directed beta-hairpin fold.
Febs Open Bio, 15, 2025
|
|
4URQ
 
 | | Crystal Structure of GGDEF domain (I site mutant) from T.maritima | | Descriptor: | DIGUANYLATE CYCLASE | | Authors: | Deepthi, A, Liew, C.W, Liang, Z.X, Swamianthan, K, Lescar, J. | | Deposit date: | 2014-07-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Diguanylate Cyclase from Thermotoga Maritima: Insights Into Activation, Feedback Inhibition and Thermostability
Plos One, 9, 2014
|
|
1CSS
 
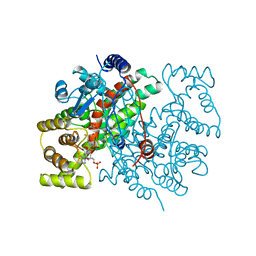 | |
4V07
 
 | | Dimeric pseudorabies virus protease pUL26N at 2.1 A resolution | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, UL26 | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-09-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease
Plos Pathog., 11, 2015
|
|
3NPZ
 
 | | Prolactin Receptor (PRLR) Complexed with the Natural Hormone (PRL) | | Descriptor: | Prolactin, Prolactin receptor | | Authors: | Van Agthoven, J, England, P, Goffin, V, Broutin, I. | | Deposit date: | 2010-06-29 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural characterization of the stem-stem dimerization interface between prolactin receptor chains complexed with the natural hormone.
J.Mol.Biol., 404, 2010
|
|
