3G0T
 
 | |
7ALK
 
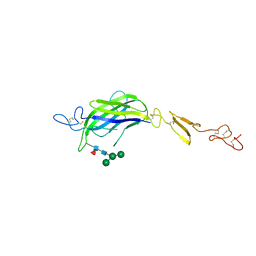 | | Structure of Drosophila C2-DSL-EGF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neurogenic locus protein delta, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
7ALT
 
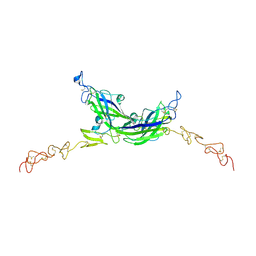 | | Structure of Drosophila Serrate C2-DSL-EGF1-EGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
3FSV
 
 | |
3FT0
 
 | |
3FSW
 
 | |
3FSZ
 
 | |
3FSA
 
 | |
3FS9
 
 | |
3E1I
 
 | |
1UIO
 
 | | ADENOSINE DEAMINASE (HIS 238 ALA MUTANT) | | Descriptor: | 6-HYDROXY-7,8-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-08-30 | | Release date: | 1997-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
2OQJ
 
 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
3C15
 
 | |
1UPM
 
 | | ACTIVATED SPINACH RUBISCO COMPLEXED WITH 2-CARBOXYARABINITOL 2 BISPHOSPHAT AND CA2+. | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Andersson, I. | | Deposit date: | 2003-10-08 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Supports Loop Closure But not Catalysis in Rubisco
J.Mol.Biol., 334, 2003
|
|
3C16
 
 | |
1UPP
 
 | | SPINACH RUBISCO IN COMPLEX WITH 2-CARBOXYARABINITOL 2 BISPHOSPHATE and Calcium. | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN, ... | | Authors: | Karkehabadi, S, Taylor, T.C, Andersson, I. | | Deposit date: | 2003-10-09 | | Release date: | 2003-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Calcium Supports Loop Closure But not Catalysis in Rubisco
J.Mol.Biol., 334, 2003
|
|
3C14
 
 | |
3CX8
 
 | |
1UIP
 
 | | ADENOSINE DEAMINASE (HIS 238 GLU MUTANT) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-08-30 | | Release date: | 1997-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
2K6A
 
 | | Solution structure of EAS D15 truncation mutant | | Descriptor: | Hydrophobin | | Authors: | Kwan, A.H. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The Cys3-Cys4 loop of the hydrophobin EAS is not required for rodlet formation and surface activity.
J.Mol.Biol., 382, 2008
|
|
1G86
 
 | | CHARCOT-LEYDEN CRYSTAL PROTEIN/N-ETHYLMALEIMIDE COMPLEX | | Descriptor: | CHARCOT-LEYDEN CRYSTAL PROTEIN, N-ETHYLMALEIMIDE | | Authors: | Ackerman, S.J, Liu, L, Kwatia, M.A, Savage, M.P, Leonidas, D.D, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2000-11-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Charcot-Leyden crystal protein (galectin-10) is not a dual function galectin with lysophospholipase activity but binds a lysophospholipase inhibitor in a novel structural fashion.
J.Biol.Chem., 277, 2002
|
|
5A2W
 
 | | Crystal structure of mtPAP in complex with ATPgammaS | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, MITOCHONDRIAL PROTEIN, ... | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
4WP0
 
 | | Crystal structure of human kynurenine aminotransferase-I with a C-terminal V5-hexahistidine tag | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase-I in a novel space group
To be published
|
|
3G7D
 
 | | Native PhpD with Cadmium Atoms | | Descriptor: | CADMIUM ION, PhpD | | Authors: | Nair, S.K. | | Deposit date: | 2009-02-09 | | Release date: | 2009-06-09 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An unusual carbon-carbon bond cleavage reaction during phosphinothricin biosynthesis.
Nature, 459, 2009
|
|
3BBF
 
 | | Crystal structure of the NM23-H2 transcription factor complex with GDP | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Weichsel, A, Montfort, W.R. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III1.
Mol.Cancer Ther., 8, 2009
|
|
