1MV0
 
 | |
1IGY
 
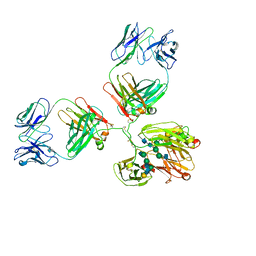 | | STRUCTURE OF IMMUNOGLOBULIN | | Descriptor: | IGG1 INTACT ANTIBODY MAB61.1.3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Harris, L.J, McPherson, A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystallographic structure of an intact IgG1 monoclonal antibody.
J.Mol.Biol., 275, 1998
|
|
1IKD
 
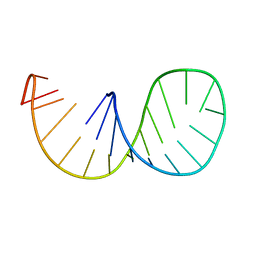 | | ACCEPTOR STEM, NMR, 30 STRUCTURES | | Descriptor: | TRNA ALA ACCEPTOR STEM | | Authors: | Ramos, A, Varani, G. | | Deposit date: | 1996-11-15 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the acceptor stem of Escherichia coli tRNA Ala: role of the G3.U70 base pair in synthetase recognition.
Nucleic Acids Res., 25, 1997
|
|
1NAN
 
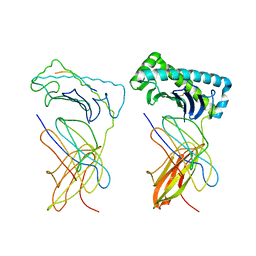 | | MCH CLASS I H-2KB MOLECULE COMPLEXED WITH PBM1 PEPTIDE | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
1IFP
 
 | |
1JF3
 
 | | Crystal Structure Of Component III Glycera Dibranchiata Monomeric Hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, monomer hemoglobin component III | | Authors: | Park, H.J, Yang, C, Treff, N, Satterlee, J.D, Kang, C.H. | | Deposit date: | 2001-06-20 | | Release date: | 2002-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Unligated and CN-Ligated Glycera dibranchiata Monomer
Ferric Hemoglobin Components III and IV
Proteins, 49, 2002
|
|
1NAM
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
1N7P
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W292A/F343V Double Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7Q
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W291A/W292A Double Mutant complex with hyaluronan hexasacchride | | Descriptor: | HYALURONIDASE, beta-D-galactopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1N7N
 
 | | Streptococcus pneumoniae Hyaluronate Lyase W292A Mutant | | Descriptor: | HYALURONIDASE | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
1NE6
 
 | | Crystal structure of Sp-cAMP binding R1a subunit of cAMP-dependent protein kinase | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, cAMP-dependent protein kinase type I-alpha regulatory chain | | Authors: | Wu, J, Jones, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2002-12-10 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of RIalpha Subunit of Cyclic Adenosine 5'-Monophosphate (cAMP)-Dependent Protein Kinase Complexed with (R(p))-Adenosine 3',5'-Cyclic Monophosphothioate and (S(p))-Adenosine 3',5'-Cyclic Monophosphothioate, the Phosphothioate Analogues of cAMP.
Biochemistry, 43, 2004
|
|
1N7O
 
 | | Streptococcus pneumoniae Hyaluronate Lyase F343V Mutant | | Descriptor: | hyaluronidase | | Authors: | Nukui, M, Taylor, K.B, McPherson, D.T, Shigenaga, M, Jedrzejas, M.J. | | Deposit date: | 2002-11-16 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The function of hydrophobic residues in the catalytic cleft of Streptococcus
pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms
J.Biol.Chem., 278, 2003
|
|
5M06
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Serine/threonine-protein kinase PknI | | Authors: | Wagner, T, Lisa, M.N, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5M07
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain, C20A mutant | | Descriptor: | SODIUM ION, Serine/threonine-protein kinase PknI | | Authors: | Lisa, M.N, Wagner, T, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5M08
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain, C20A_R136A double mutant | | Descriptor: | Serine/threonine-protein kinase PknI | | Authors: | Lisa, M.N, Wagner, T, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5M09
 
 | | Crystal structure of Mycobacterium tuberculosis PknI kinase domain, C20A_R136N double mutant | | Descriptor: | SODIUM ION, Serine/threonine-protein kinase PknI | | Authors: | Lisa, M.N, Wagner, T, Alexandre, M, Barilone, N, Raynal, B, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The crystal structure of PknI from Mycobacterium tuberculosis shows an inactive, pseudokinase-like conformation.
FEBS J., 284, 2017
|
|
5OCA
 
 | | PCSK9:Fab Complex with Dextran Sulfate | | Descriptor: | 2,3,4-tri-O-sulfo-beta-D-altropyranose-(1-6)-2,3-di-O-sulfo-alpha-L-glucopyranose, Fab from LDLR competitive antibody: Heavy chain, Fab from LDLR competitive antibody: Light chain, ... | | Authors: | Thirup, S.S, Vilstrup, J.P. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Heparan sulfate proteoglycans present PCSK9 to the LDL receptor.
Nat Commun, 8, 2017
|
|
3UNY
 
 | | Bacillus cereus phosphopentomutase T85E variant soaked with glucose 1,6-bisphosphate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3UQC
 
 | |
5W8M
 
 | | Crystal structure of Chaetomium thermophilum Vps29 | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, Vacuolar protein sorting-associated protein 29 | | Authors: | Collins, B.M, Leneva, N. | | Deposit date: | 2017-06-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the membrane-assembled retromer coat determined by cryo-electron tomography.
Nature, 561, 2018
|
|
5L7K
 
 | |
6VQX
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PPN
 
 | | Structure of S. pombe Lsm2-8 with unprocessed U6 snRNA | | Descriptor: | Mimic of unprocessed U6 snRNA, Probable U6 snRNA-associated Sm-like protein LSm3, Probable U6 snRNA-associated Sm-like protein LSm4, ... | | Authors: | Montemayor, E.J, Butcher, S.E. | | Deposit date: | 2019-07-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular basis for the distinct cellular functions of the Lsm1-7 and Lsm2-8 complexes.
Rna, 26, 2020
|
|
6VQW
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3DD7
 
 | |
