6HSU
 
 | |
8U6M
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((6-chloro-2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ751), a non-nucleoside inhibitor | | Descriptor: | N-[2-(2-{[(4R)-6-chloro-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy)ethyl)-N-methylacrylamide (JLJ736), a non-nucleoside inhibitor | | Descriptor: | N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8U6S
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 8-(2-(3-morpholino-3-oxopropoxy)phenoxy)indolizine-2-carbonitrile (JLJ757), a non-nucleoside inhibitor | | Descriptor: | (4S)-8-{2-[3-(morpholin-4-yl)-3-oxopropoxy]phenoxy}indolizine-2-carbonitrile, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
7NL4
 
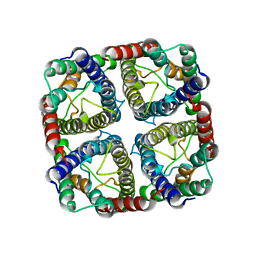 | | OsNIP2;1 silicon transporter from rice | | Descriptor: | Aquaporin NIP2-1, CADMIUM ION | | Authors: | van den Berg, B. | | Deposit date: | 2021-02-22 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Silicic Acid Uptake by Higher Plants.
J.Mol.Biol., 433, 2021
|
|
8U6C
 
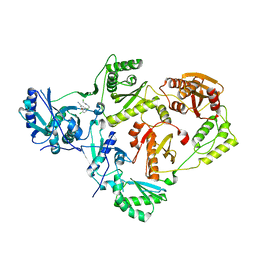 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 2-chloro-N-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenethyl)acetamide (JLJ732), a non-nucleoside inhibitor | | Descriptor: | 2-chloro-N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenyl]ethyl}acetamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
7N13
 
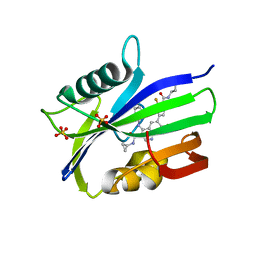 | | Crystal structure of MTH1 in complex with compound 32 | | Descriptor: | 4-anilino-6-[4-(butylcarbamoyl)-3-fluorophenyl]-N-cyclopropyl-7-fluoroquinoline-3-carboxamide, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eron, S.J. | | Deposit date: | 2021-05-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of an AchillesTAG degradation system and its application to control CAR-T activity
Curr Res Chem Biol, 1, 2021
|
|
8BJW
 
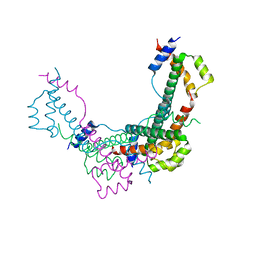 | |
6H88
 
 | | MamM CTD D249N | | Descriptor: | BETA-MERCAPTOETHANOL, Magnetosome protein MamM, Cation efflux protein family | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MamM CTD D249N
To Be Published
|
|
8U6R
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((2-cyanoindolizin-8-yl)oxy)phenoxy)-N-(2,2-difluoroethyl)propanamide (JLJ756), a non-nucleoside inhibitor | | Descriptor: | 3-(2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)-N-(2,2-difluoroethyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8UPI
 
 | | Structure of a periplasmic peptide binding protein from Mesorhizobium sp. AP09 bound to aminoserine | | Descriptor: | 1,2-ETHANEDIOL, AMINOSERINE, CALCIUM ION, ... | | Authors: | Frkic, R.L, Smith, O.B, Rahman, M, Kaczmarski, J.A, Jackson, C.J. | | Deposit date: | 2023-10-22 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Characterization of a Bacterial Periplasmic Solute Binding Protein That Binds l-Amino Acid Amides.
Biochemistry, 63, 2024
|
|
6E3M
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 6-(3-carboxyphenyl)-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-carboxyphenyl)-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
8B5K
 
 | | Structure of haloalkane dehalogenase DmmarA from Mycobacterium marinum at pH 6.5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Snajdarova, K, Marek, M. | | Deposit date: | 2022-09-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Atypical homodimerization revealed by the structure of the (S)-enantioselective haloalkane dehalogenase DmmarA from Mycobacterium marinum.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8UVZ
 
 | |
6HHZ
 
 | | Klebsiella pneumoniae Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)cytidine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-seryl)-sulfamoyl)cytidine, CALCIUM ION, ... | | Authors: | Pang, L, De Graef, S, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2018-08-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
8BMY
 
 | | Bacteroides thetaiotaomicron surface lipoprotein bound to cyanocobalamin | | Descriptor: | CHLORIDE ION, CYANOCOBALAMIN, Putative surface layer protein, ... | | Authors: | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2022-11-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
6HIG
 
 | | hPD-1/NBO1a Fab complex | | Descriptor: | Heavy Chain, Light Chain, Programmed cell death protein 1 | | Authors: | Loredo-Varela, J.L, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tumor suppression of novel anti-PD-1 antibodies mediated through CD28 costimulatory pathway.
J.Exp.Med., 216, 2019
|
|
8U6H
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
6HMW
 
 | | Cholera toxin classical B-pentamer in complex with fucose | | Descriptor: | BICINE, CALCIUM ION, Cholera enterotoxin B-subunit, ... | | Authors: | Krengel, U, Heim, J.B. | | Deposit date: | 2018-09-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of cholera toxin in complex with fucosylated receptors point to importance of secondary binding site.
Sci Rep, 9, 2019
|
|
6ES8
 
 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag protein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2017-10-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
8BJD
 
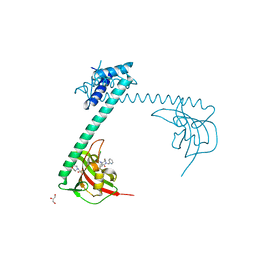 | | Full length structure of LpMIP with bound inhibitor JK095 | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-04 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BK4
 
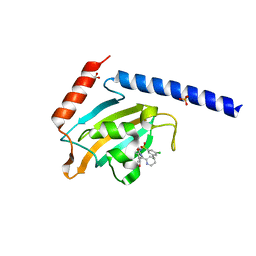 | | Full length structure of the apo-state LpMIP. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-[(1~{S})-1-pyridin-2-ylethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, GLYCEROL, Macrophage infectivity potentiator, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
8BK5
 
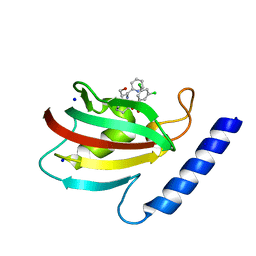 | | A structure of the truncated LpMIP with bound inhibitor JK095. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase, SODIUM ION | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
6ESC
 
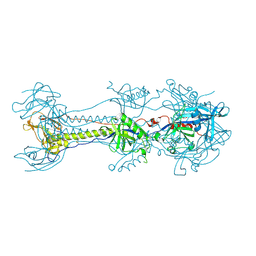 | | Crystal structure of Pseudorabies virus glycoprotein B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, GLYCEROL | | Authors: | Backovic, M, Vaney, M.C, Rey, F.A, Haouz, A. | | Deposit date: | 2017-10-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Function Dissection of Pseudorabies Virus Glycoprotein B Fusion Loops.
J.Virol., 92, 2018
|
|
8BN0
 
 | | Bacteroides thetaiotaomicron surface protein BT1954 bound to | | Descriptor: | CHLORIDE ION, COB(II)INAMIDE, CYANIDE ION, ... | | Authors: | Abellon-Ruiz, J, Jana, K, Silale, A, Basle, A, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2022-11-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B 12 uptake in gut Bacteroides.
Nat Commun, 14, 2023
|
|
