5B4O
 
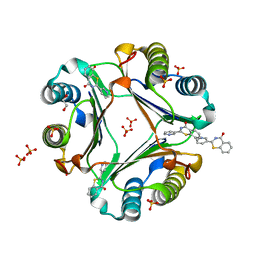 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
1DFO
 
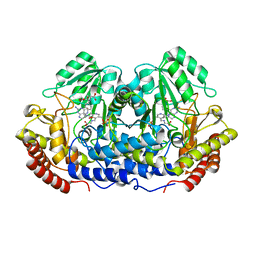 | | CRYSTAL STRUCTURE AT 2.4 ANGSTROM RESOLUTION OF E. COLI SERINE HYDROXYMETHYLTRANSFERASE IN COMPLEX WITH GLYCINE AND 5-FORMYL TETRAHYDROFOLATE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Scarsdale, J.N, Radaev, S, Kazanina, G, Schirch, V, Wright, H.T. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate.
J.Mol.Biol., 296, 2000
|
|
6DHK
 
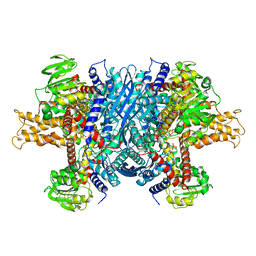 | | Bovine glutamate dehydrogenase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation.
Biochemistry, 42, 2003
|
|
1ON3
 
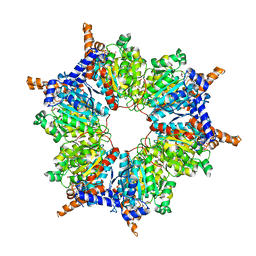 | | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with methylmalonyl-coenzyme a and methylmalonic acid bound) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CADMIUM ION, METHYLMALONIC ACID, ... | | Authors: | Hall, P.R, Wang, Y.-F, Rivera-Hainaj, R.E, Zheng, X, Pustai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core
Embo J., 22, 2003
|
|
3B9D
 
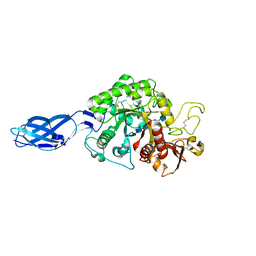 | | Crystal structure of Vibrio harveyi chitinase A complexed with pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A | | Authors: | Songsiriritthigul, C, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
3B9E
 
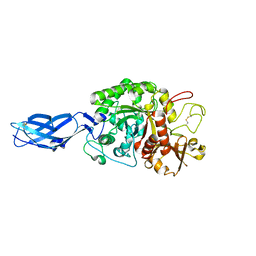 | | Crystal structure of inactive mutant E315M chitinase A from Vibrio harveyi | | Descriptor: | Chitinase A | | Authors: | Songsiriritthigul, C, Pantoom, S, Aguda, A.H, Robinson, R.C, Suginta, W. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Vibrio harveyi chitinase A complexed with chitooligosaccharides: implications for the catalytic mechanism
J.Struct.Biol., 162, 2008
|
|
2BHP
 
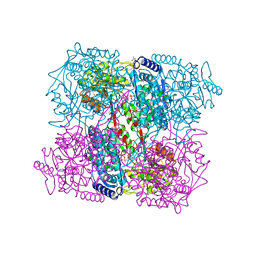 | | Crystal Analysis of 1-Pyrroline-5-Carboxylate Dehydrogenase from Thermus with bound NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Tahirov, T.H. | | Deposit date: | 2005-01-16 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
1LZV
 
 | | Site-Specific Mutant (Tyr7 replaced with His) of Human Carbonic Anhydrase II | | Descriptor: | Carbonic Anhydrase II, ZINC ION | | Authors: | Tu, C.K, Qian, M, An, H, Wadhwa, N.R, Duda, D.M, Yoshioka, C, Pathak, Y, McKenna, R, Laipis, P.J, Silverman, D.N. | | Deposit date: | 2002-06-11 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic analysis of multiple proton shuttles in the active site of human carbonic anhydrase.
J.Biol.Chem., 277, 2002
|
|
1DQK
 
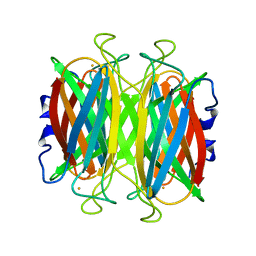 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE IN THE REDUCED STATE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
5VOU
 
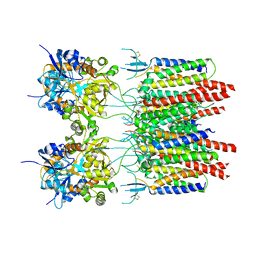 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Chen, S, Zhao, Y, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
5OKS
 
 | | Non-conservatively refined structure of Gan1D, a 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with 6-phospho-beta-glucose | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
5JFD
 
 | | Thrombin in complex with (S)-N-(2-(aminomethyl)-5-chlorobenzyl)-1-((benzylsulfonyl)-D-arginyl)pyrrolidine-2-carboxamide | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloro-phenyl]methyl]-1-[(2R)-5-carbamimidamido-2-(phenylmethylsulfonylamino)pentanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
3A05
 
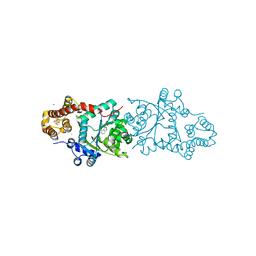 | | Crystal structure of tryptophanyl-tRNA synthetase from hyperthermophilic archaeon, Aeropyrum pernix K1 complex with tryptophan | | Descriptor: | CADMIUM ION, IRON/SULFUR CLUSTER, TRYPTOPHAN, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Hasegawa, T. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tryptophanyl-tRNA synthetase from hyperthermophilic archaeon, Aeropyrum pernix K1
To be Published
|
|
3T4K
 
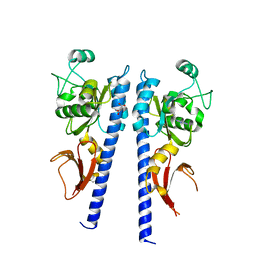 | |
2VSS
 
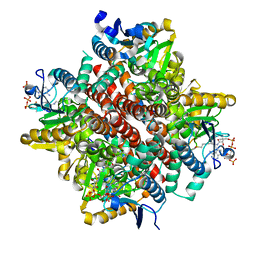 | | Wild-type Hydroxycinnamoyl-CoA hydratase lyase in complex with acetyl- CoA and vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
1MTB
 
 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
1N1L
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND INHIBITOR (GW472467X) | | Descriptor: | HCV NS3 SERINE PROTEASE, NS4A COFACTOR, ZINC ION, ... | | Authors: | Andrews, D.M, Chaignot, H, Coomber, B.A, Good, A.C, Hind, S.L, Jones, P.S, Mill, G, Robinson, J.E, Skarzynski, T, Slater, M.J, Somers, D.O.N. | | Deposit date: | 2002-10-18 | | Release date: | 2003-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 2. The use of X-ray Crystal Structure Data in the Optimisation of P3 and P4 Substituents
Org.Lett., 4, 2002
|
|
1MT7
 
 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | ACETATE ION, PROTEASE RETROPEPSIN, Substrate analogue | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
3NIG
 
 | | The Closed Headpiece of Integrin IIb 3 and its Complex with an IIb 3 -Specific Antagonist That Does Not Induce Opening | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J.H, Zhu, J.Q, Springer, T.A. | | Deposit date: | 2010-06-15 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Closed headpiece of integrin {alpha}IIb{beta}3 and its complex with an {alpha}IIb{beta}3-specific antagonist that does not induce opening.
Blood, 116, 2010
|
|
1M9P
 
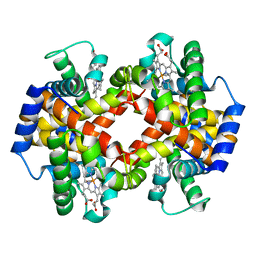 | | Crystalline Human Carbonmonoxy Hemoglobin C Exhibits The R2 Quaternary State at Neutral pH In The Presence of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3C8Z
 
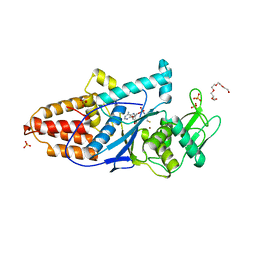 | | The 1.6 A Crystal Structure of MshC: The Rate Limiting Enzyme in the Mycothiol Biosynthetic Pathway | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Cysteinyl-tRNA synthetase, ... | | Authors: | Tremblay, L.W, Fan, F, Vetting, M.W, Blanchard, J.S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway.
Biochemistry, 47, 2008
|
|
1JBW
 
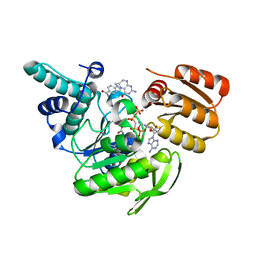 | | FPGS-AMPPCP-folate complex | | Descriptor: | 5,10-METHYLENE-6-HYDROFOLIC ACID, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, FOLYLPOLYGLUTAMATE SYNTHASE, ... | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
2QKT
 
 | |
1YI9
 
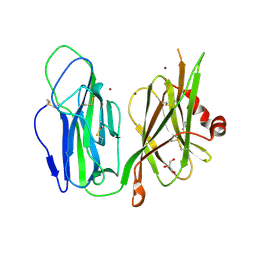 | | Crystal Structure Analysis of the oxidized form of the M314I mutant of Peptidylglycine alpha-Hydroxylating Monooxygenase | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Siebert, X, Eipper, B.A, Mains, R.E, Prigge, S.T, Blackburn, N.J, Amzel, L.M. | | Deposit date: | 2005-01-11 | | Release date: | 2005-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic copper of Peptidylglycine alpha-Hydroxylating Monooxygenase also plays a critical structural role.
Biophys.J., 89, 2005
|
|
3BQJ
 
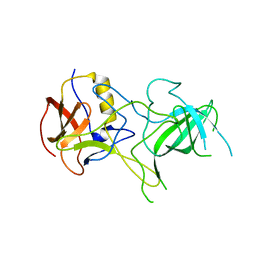 | | VA387 polypeptide | | Descriptor: | va387 polypeptide | | Authors: | Bu, W, Mamedova, A, Tan, M, Jiang, J, Hegde, R. | | Deposit date: | 2007-12-20 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
