5OXF
 
 | |
5OWV
 
 | |
3R45
 
 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
3RJS
 
 | |
3Q5W
 
 | |
3Q5X
 
 | | Structure of proteasome tether | | Descriptor: | Protein cut8 | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-12-30 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of Proteasome Tether
To be Published
|
|
4G2L
 
 | | Human PDE9 in complex with selective compound | | Descriptor: | 1-cyclopentyl-6-{(1R)-1-[3-(pyrimidin-2-yl)azetidin-1-yl]ethyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-07-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
4DDV
 
 | |
4DDT
 
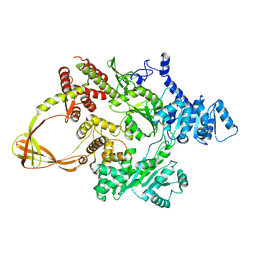 | | Thermotoga maritima reverse gyrase, C2 FORM 2 | | Descriptor: | Reverse gyrase, ZINC ION | | Authors: | Rudolph, M.G, Klostermeier, D. | | Deposit date: | 2012-01-19 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of Thermotoga maritima reverse gyrase: inferences for the mechanism of positive DNA supercoiling.
Nucleic Acids Res., 41, 2013
|
|
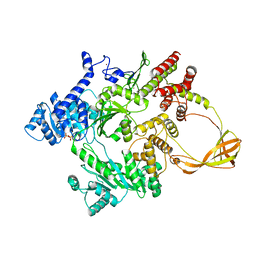
4DDW
 
 | |
4E90
 
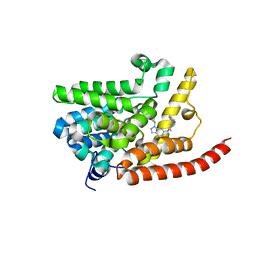 | | Human phosphodiesterase 9 in complex with inhibitors | | Descriptor: | 6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-03-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
4FAB
 
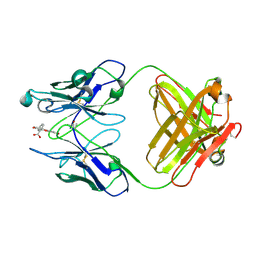 | | THREE-DIMENSIONAL STRUCTURE OF A FLUORESCEIN-FAB COMPLEX CRYSTALLIZED IN 2-METHYL-2,4-PENTANEDIOL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, IGG2A-KAPPA 4-4-20 FAB (HEAVY CHAIN), ... | | Authors: | Herron, J.N, He, X, Mason, M.L, Vossjunior, E.W, Edmundson, A.B. | | Deposit date: | 1989-04-10 | | Release date: | 1990-07-15 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of a fluorescein-Fab complex crystallized in 2-methyl-2,4-pentanediol.
Proteins, 5, 1989
|
|
4E89
 
 | | Crystal Structure of RnaseH from gammaretrovirus | | Descriptor: | CADMIUM ION, MAGNESIUM ION, RNase H | | Authors: | Kim, J.H, Kim, S.J. | | Deposit date: | 2012-03-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of xenotropic murine leukaemia virus-related virus (XMRV) ribonuclease H
Biosci.Rep., 32, 2012
|
|
4H8S
 
 | | Crystal structure of human APPL2BARPH domain | | Descriptor: | DCC-interacting protein 13-beta | | Authors: | Martin, J.L, King, G.J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Membrane Curvature Protein Exhibits Interdomain Flexibility and Binds a Small GTPase.
J.Biol.Chem., 287, 2012
|
|
4HNK
 
 | | Crystal structure of an Enzyme | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL | | Authors: | El Bakkouri, M, Calmettes, C, Wernimont, A.K, Houry, W.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-19 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Inactive Subunit of the Apicoplast-localized Caseinolytic Protease Complex of Plasmodium falciparum.
J.Biol.Chem., 288, 2013
|
|
4G2J
 
 | | Human pde9 in complex with selective compound | | Descriptor: | 1-cyclopentyl-6-[(1R)-1-(3-phenoxyazetidin-1-yl)ethyl]-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-07-12 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
5MT9
 
 | | Human insulin in complex with serotonin and arginine | | Descriptor: | ARGININE, CHLORIDE ION, Insulin, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2017-01-07 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
5MT3
 
 | | Human insulin in complex with serotonin and arginine | | Descriptor: | ARGININE, CHLORIDE ION, Insulin, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2017-01-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
5NJP
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJQ
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NV8
 
 | | Structural basis for EarP-mediated arginine glycosylation of translation elongation factor EF-P | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, EF-P arginine 32 rhamnosyl-transferase | | Authors: | Macosek, J, Krafczyk, R, Jagtap, P.K.A, Lassaka, J, Hennig, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Basis for EarP-Mediated Arginine Glycosylation of Translation Elongation Factor EF-P.
MBio, 8, 2017
|
|
5NU5
 
 | |
5NRW
 
 | |
5NU3
 
 | | Crystal structure of the human bromodomain of CREBBP bound to the inhibitor XDM-CBP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CREB-binding protein, SULFATE ION, ... | | Authors: | Huegle, M, Wohlwend, D. | | Deposit date: | 2017-04-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Beyond the BET Family: Targeting CBP/p300 with 4-Acyl Pyrroles.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MAM
 
 | | Human insulin in complex with serotonin | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2016-11-03 | | Release date: | 2017-04-05 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Computational and structural evidence for neurotransmitter-mediated modulation of the oligomeric states of human insulin in storage granules.
J. Biol. Chem., 292, 2017
|
|
