6DZQ
 
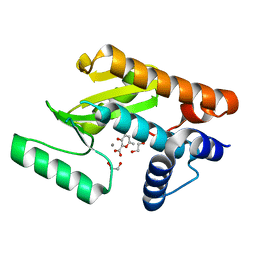 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Cohen, S.M. | | Deposit date: | 2018-07-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease.
J. Med. Chem., 61, 2018
|
|
8RCB
 
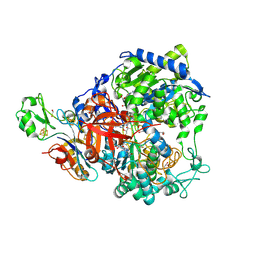 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air (in a not degassed drop) for 34 min in the presence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RCC
 
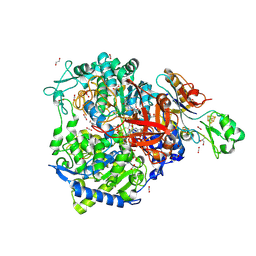 | | W-formate dehydrogenase from Desulfovibrio vulgaris - aerobic soaked with 48 bar CO2 for 1 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBON DIOXIDE, ... | | Authors: | Vilela-Alves, G, Carpentier, P, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8BDM
 
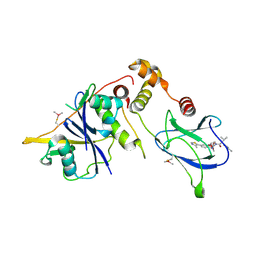 | | VCB in complex with compound 26 | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-(2-methoxyethoxy)-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{R})-3-methyl-2-(3-methyl-1,2-oxazol-5-yl)butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8RXG
 
 | |
8RYU
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 10 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [6.9 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8BDW
 
 | | Crystal structure of CnaB2 domain from Lactobacillus plantarum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell surface adherence protein,collagen-binding domain, LPXTG-motif cell wall anchor, ... | | Authors: | Taberman, H, Hakanpaa, J, Linder, M.B, Aranko, A.S. | | Deposit date: | 2022-10-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Biomolecular Click Reactions Using a Minimal pH-Activated Catcher/Tag Pair for Producing Native-Sized Spider-Silk Proteins.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6E0L
 
 | | Structure of Rhodothermus marinus CdnE c-UMP-AMP synthase with Apcpp and Upnpp | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
7ML8
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ001023038 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-phenyl-1,6-dihydropyrimidine-4-carboxamide, GLUTAMIC ACID, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | Deposit date: | 2021-04-27 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with St Jude compound AC067-19
To Be Published
|
|
8RQJ
 
 | |
8BDI
 
 | | VCB in complex with compound 32 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-3-(methylamino)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-3-oxidanylidene-propyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8RFT
 
 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 17 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [10.37 MGy] - water ligand + decarboxylated AspCAT | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7M4S
 
 | |
8RGD
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 15 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [10.2 MGy] | | Descriptor: | CARBON DIOXIDE, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
7MK1
 
 | | Structure of a protein-modified aptamer complex | | Descriptor: | Antiviral innate immune response receptor RIG-I, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Ren, X, Pyle, A.M. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving A RIG-I Antagonist: A Modified DNA Aptamer Mimics Viral RNA.
J.Mol.Biol., 433, 2021
|
|
8RC8
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Exposed to air for 0 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8RFW
 
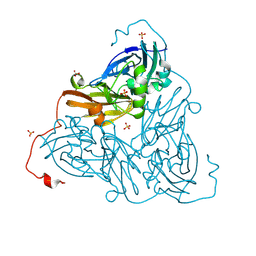 | | High pH (8.0) as-isolated MSOX movie series dataset 2 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [0.82 MGy] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
8S2Q
 
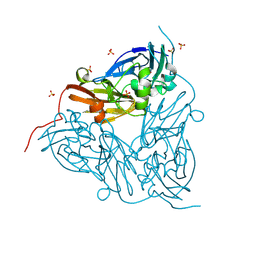 | | High pH (8.0) as-isolated MSOX movie series dataset 2 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [0.7 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
6CAC
 
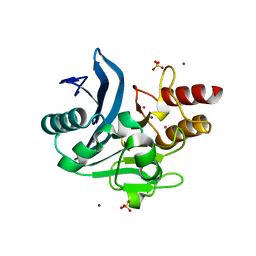 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
8RZI
 
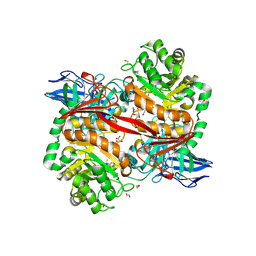 | | ZgGH129 from Zobellia galactanivorans soaked with 1,2-diF-ADG (3,6-Anhydro-2-deoxy-2-fluoro-a-D-galactopyranosyl fluoride) resulting in a trapped glycosyl-enzyme intermediate. | | Descriptor: | (1~{R},4~{S},5~{S},8~{S})-4-fluoranyl-2,6-dioxabicyclo[3.2.1]octan-8-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8BDN
 
 | | VCB in complex with compound 23 | | Descriptor: | (2~{R})-3-methyl-1-[(2~{S},4~{R})-2-[(5~{R})-5-methyl-5-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-4~{H}-1,2,4-oxadiazol-3-yl]-4-oxidanyl-pyrrolidin-1-yl]-2-(3-methyl-1,2-oxazol-5-yl)butan-1-one, Elongin-B, Elongin-C, ... | | Authors: | Sorrell, F.J, Mueller, J.E, Lehmann, M, Wegener, A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Systematic Potency and Property Assessment of VHL Ligands and Implications on PROTAC Design.
Chemmedchem, 18, 2023
|
|
8RFU
 
 | | Low pH (5.5) nitrite-bound MSOX movie series dataset 40 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [24.4 MGy] - water ligand + decarboxylated AspCAT (final) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
6JTP
 
 | | Crystal structure of HLA-C08 in complex with a tumor mut9m peptide | | Descriptor: | 9-mer peptide, Beta-2-microglobulin, HLA class I antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Lei, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immune-based mutation classification enables neoantigen prioritization and immune feature discovery in cancer immunotherapy.
Oncoimmunology, 10, 2021
|
|
8RCA
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air for 1 h in the absence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
8BF1
 
 | |
