1KBP
 
 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-02-20 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
1FXT
 
 | | STRUCTURE OF A CONJUGATING ENZYME-UBIQUITIN THIOLESTER COMPLEX | | Descriptor: | UBIQUITIN, UBIQUITIN-CONJUGATING ENZYME E2-24 KDA | | Authors: | Hamilton, K.S, Shaw, G.S, Williams, R.S, Huzil, J.T, McKenna, S, Ptak, C, Glover, M, Ellison, M.J. | | Deposit date: | 2000-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail.
Structure, 9, 2001
|
|
1KMY
 
 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 2,3-dihydroxybiphenyl under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
2P73
 
 | | crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Y, Xiang, Y, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
4HHF
 
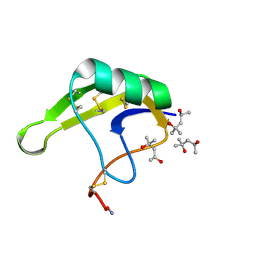 | | Crystal Structure of chemically synthesized scorpion alpha-toxin OD1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-toxin OD1 | | Authors: | Wang, C.I.A, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-10-09 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical engineering and structural and pharmacological characterization of the alpha-scorpion toxin OD1.
Acs Chem.Biol., 8, 2013
|
|
1YAZ
 
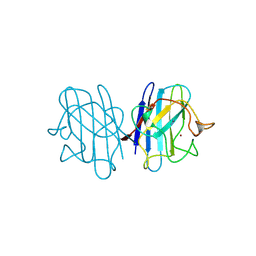 | | AZIDE-BOUND YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | AZIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
3PFV
 
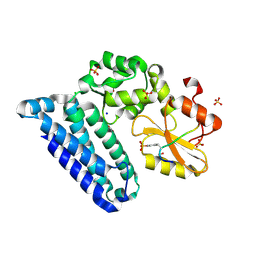 | | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide | | Descriptor: | 1,2-ETHANEDIOL, 11-meric peptide from Epidermal growth factor receptor, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Guo, K, Cooper, C.D.O, Ayinampudi, V, Krojer, T, Muniz, J.R.C, Vollmar, M, Canning, P, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Cbl-b TKB domain in complex with EGFR pY1069 peptide
To be Published
|
|
4LHK
 
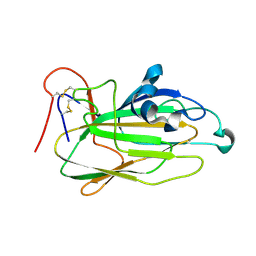 | |
1KOK
 
 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (MpCcP) | | Descriptor: | Cytochrome c Peroxidase, FE(III)-(4-MESOPORPHYRINONE) | | Authors: | Bhaskar, B, Immoos, C.E, Cohen, M.S, Barrows, T.P, Farmer, P.J, Poulos, T.L. | | Deposit date: | 2001-12-20 | | Release date: | 2002-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mesopone cytochrome c peroxidase: functional model of heme oxygenated oxidases.
J.Inorg.Biochem., 91, 2002
|
|
3OKY
 
 | | Plexin A2 in complex with Semaphorin 6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Janssen, B.J.C, Robinson, R.A, Bell, C.H, Siebold, C, Jones, E.Y. | | Deposit date: | 2010-08-25 | | Release date: | 2010-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural basis of semaphorin-plexin signalling.
Nature, 467, 2010
|
|
2HU7
 
 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | ACETYL GROUP, Acylamino-acid-releasing enzyme, GLYCEROL, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HU8
 
 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | 2-AMINOBENZOIC ACID, Acylamino-acid-releasing enzyme, GLYCINE, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HU5
 
 | | Binding of inhibitors by Acylaminoacyl-peptidase | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, GLYCINE, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
8CL9
 
 | | Tubulin-DARPin D1 complex | | Descriptor: | DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wranik, M, Bertrand, Q, Weinert, T, Kepa, M.W, Steinmetz, M, Standfuss, J. | | Deposit date: | 2023-02-16 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-reservoir extruder for time-resolved serial protein crystallography and compound screening at X-ray free-electron lasers.
Nat Commun, 14, 2023
|
|
3KBS
 
 | |
3KBV
 
 | |
5QBY
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-[2-chloro-5-(1-{3-[4-(6-chloro-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-1-yl)piperidin-1-yl]propyl}-6-oxo-1,6-dihydropyrimidin-5-yl)benzyl]-4-fluorobenzamide | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diazinones as P2 replacements for pyrazole-based cathepsin S inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7F6W
 
 | | Crystal structure of Saccharomyces cerevisiae lysyl-tRNA Synthetase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysine--tRNA ligase | | Authors: | Wu, S, Li, P, Hei, Z, Zheng, L, Wang, J, Fang, P. | | Deposit date: | 2021-06-26 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Human lysyl-tRNA synthetase evolves a dynamic structure that can be stabilized by forming complex.
Cell.Mol.Life Sci., 79, 2022
|
|
6EKE
 
 | |
5KBK
 
 | | Candida Albicans Superoxide Dismutase 5 (SOD5), E110A Mutant | | Descriptor: | COPPER (I) ION, Cell surface Cu-only superoxide dismutase 5, SULFATE ION | | Authors: | Galaleldeen, A, Peterson, R.L, Villarreal, J, Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | The Phylogeny and Active Site Design of Eukaryotic Copper-only Superoxide Dismutases.
J.Biol.Chem., 291, 2016
|
|
5QC4
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-[5-[5-ethanoyl-1-[(2~{R})-2-oxidanyl-3-[4-(2-oxidanylpropan-2-yl)piperidin-1-yl]propyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]-2-(trifluoromethyl)phenyl]sulfanyl-1-pyrrolidin-1-yl-ethanone, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thioether acetamides as P3 binding elements for tetrahydropyrido-pyrazole cathepsin S inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KBW
 
 | |
5PGV
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE | | Descriptor: | 1-(3-HYDROXYAZETIDIN-1-YL)-2-[(2S,5R)-2-(4-FLUOROPHENYL)-5-METHOXYADAMANTAN-2-YL]ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
5PGW
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3- HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE | | Descriptor: | 2-[(1R,3S,5R,7S)-2-[4-(4-FLUOROPHENYL)PHENYL]-6-HYDROXYADAMANTAN-2-YL]-1-(3-HYDROXYAZETIDIN-1-YL)ETHAN-1-ONE, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of Clinical Candidate 2-((2S,6S)-2-Phenyl-6-hydroxyadamantan-2-yl)-1-(3'-hydroxyazetidin-1-yl)ethanone [BMS-816336], an Orally Active Novel Selective 11 beta-Hydroxysteroid Dehydrogenase Type 1 Inhibitor.
J. Med. Chem., 60, 2017
|
|
