4BB4
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N-(2-methoxyethyl)-4-[(6-pyridin-4-ylquinazolin-2-yl)amino]benzamide | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf V600E Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
8D95
 
 | |
6NXL
 
 | | Ubiquitin binding variants | | Descriptor: | Polyubiquitin-B | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2F21
 
 | | human Pin1 Fip mutant | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Jager, M, Zhang, Y, Bowman, M.E, Noel, J.P, Kelly, J.W. | | Deposit date: | 2005-11-15 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function-folding relationship in a WW domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5IA2
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with compound 66 | | Descriptor: | 1,2-ETHANEDIOL, 7-(5-hydroxy-2-methylphenyl)-8-(2-methoxyphenyl)-1-methyl-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
4YTR
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4YTQ
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy D-tagatose | | Descriptor: | 1-deoxy-D-tagatose, 1-deoxy-alpha-D-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5WEY
 
 | | Joint X-ray/neutron structure of Concanavalin A with alpha1-2 D-mannobiose | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Woods, R.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Mannobiose Binding Induces Changes in Hydrogen Bonding and Protonation States of Acidic Residues in Concanavalin A As Revealed by Neutron Crystallography.
Biochemistry, 56, 2017
|
|
1BCD
 
 | |
4YW5
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
4ARV
 
 | | Yersinia kristensenii phytase apo form | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
8D7R
 
 | |
8D7H
 
 | | Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cardiotrophin-like cytokine factor 1, ... | | Authors: | Zhou, Y, Franklin, M.C. | | Deposit date: | 2022-06-07 | | Release date: | 2023-03-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly of gp130 family cytokine signaling complexes.
Sci Adv, 9, 2023
|
|
1YYD
 
 | | High Resolution Crystal Structure of Manganese Peroxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Sundaramoorthy, M, Youngs, H.L, Gold, M.H, Poulos, T.L. | | Deposit date: | 2005-02-24 | | Release date: | 2005-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Crystal Structure of Manganese Peroxidase: Substrate and Inhibitor Complexes.
Biochemistry, 44, 2005
|
|
5I34
 
 | | Adenylosuccinate synthetase from Cryptococcus neoformans complexed with GDP and IMP | | Descriptor: | Adenylosuccinate synthetase, GUANOSINE-5'-DIPHOSPHATE, INOSINIC ACID | | Authors: | Blundell, R.D, Williams, S.J, Ericsson, D, Fraser, J.A, Kobe, B. | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Disruption of de Novo Adenosine Triphosphate (ATP) Biosynthesis Abolishes Virulence in Cryptococcus neoformans.
Acs Infect Dis., 2, 2016
|
|
1AVG
 
 | |
1Z3U
 
 | |
5Z02
 
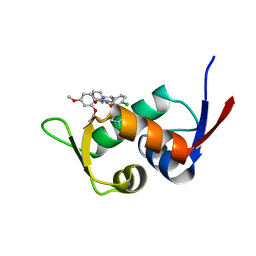 | | Crystal structure of HIS6-tagged Mdm2 with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Su, Z.D, Cheng, X.Y, Chen, R, Pi, N. | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of HIS6-tagged Mdm2 with nutlin-3a
to be published
|
|
4WP4
 
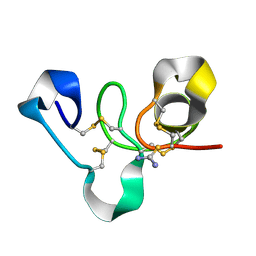 | | Hev b 6.02 (hevein) extracted from surgical gloves | | Descriptor: | Pro-hevein, THIOUREA | | Authors: | Galicia, C, Rodriguez-Romero, A. | | Deposit date: | 2014-10-17 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Impact of the vulcanization process on the structural characteristics and IgE recognition of two allergens, Hev b 2 and Hev b 6.02, extracted from latex surgical gloves.
Mol.Immunol., 65, 2015
|
|
5CKI
 
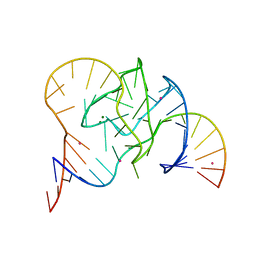 | | Crystal structure of 9DB1* deoxyribozyme (Cobalt hexammine soaked crystals) | | Descriptor: | COBALT (II) ION, DNA (44-MER), MAGNESIUM ION, ... | | Authors: | Ponce-Salvatierra, A, Hoebartner, C, Pena, V. | | Deposit date: | 2015-07-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of a DNA catalyst.
Nature, 529, 2016
|
|
8CH6
 
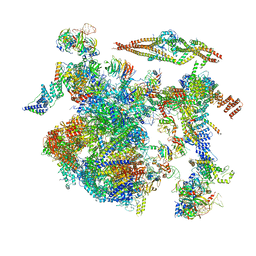 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
5MZZ
 
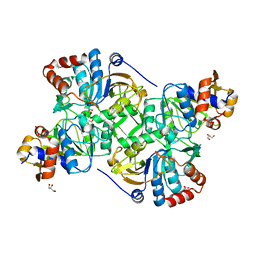 | | Crystal structure of the decarboxylase AibA/AibB in complex with 3-methylglutaconate | | Descriptor: | 3-methylpent-2-enedioic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LCQ
 
 | |
5N01
 
 | | Crystal structure of the decarboxylase AibA/AibB C56N variant | | Descriptor: | ACETATE ION, GLYCEROL, Glutaconate CoA-transferase family, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6NXK
 
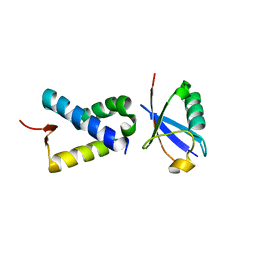 | | Ubiquitin binding variants | | Descriptor: | Anaphase-promoting complex subunit 2, Polyubiquitin-C | | Authors: | Miller, D.J, Watson, E.R. | | Deposit date: | 2019-02-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
