5T6X
 
 | | HLA-B*57:01 presenting TSTTSVASSW | | Descriptor: | Beta-2-microglobulin, Decapeptide: THR-SER-THR-THR-SER-VAL-ALA-SER-SER-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3HMV
 
 | | Catalytic domain of human phosphodiesterase 4B2B in complex with a tetrahydrobenzothiophene inhibitor | | Descriptor: | (6S)-6-methyl-2-{[(2-nitrophenyl)carbonyl]amino}-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Somers, D.O, Neu, M. | | Deposit date: | 2009-05-29 | | Release date: | 2010-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Identification of PDE4B Over 4D subtype-selective inhibitors revealing an unprecedented binding mode
Bioorg.Med.Chem., 17, 2009
|
|
5CS9
 
 | | The structure of the NK1 fragment of HGF/SF complexed with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
1E1F
 
 | | Crystal structure of a Monocot (Maize ZMGlu1) beta-glucosidase in complex with p-Nitrophenyl-beta-D-thioglucoside | | Descriptor: | 4-nitrophenyl 1-thio-beta-D-glucopyranoside, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-05-03 | | Release date: | 2001-02-19 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Monocotyledon (Maize Zmglu1) Beta-Glucosidase and a Model of its Complex with P-Nitrophenyl Beta-D-Thioglucoside
Biochem.J., 354, 2001
|
|
7ALV
 
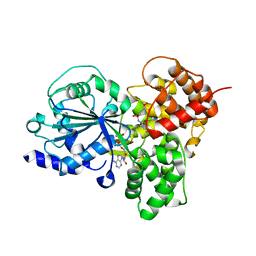 | | Crystal Structure of NLRP3 NACHT domain in complex with a potent inhibitor | | Descriptor: | 1-[4-chloranyl-2,6-di(propan-2-yl)phenyl]-3-[4-(2-oxidanylpropan-2-yl)furan-2-yl]sulfonyl-urea, ADENOSINE-5'-DIPHOSPHATE, NACHT, ... | | Authors: | Dekker, C, Hinniger, A. | | Deposit date: | 2020-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | Crystal Structure of NLRP3 NACHT Domain With an Inhibitor Defines Mechanism of Inflammasome Inhibition.
J.Mol.Biol., 433, 2021
|
|
4BCR
 
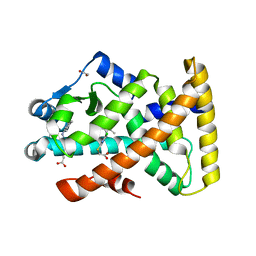 | | Structure of PPARalpha in complex with WY14643 | | Descriptor: | 1,2-ETHANEDIOL, 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR ALPHA | | Authors: | Bernardes, A, Muniz, J.R.C, Polikarpov, I. | | Deposit date: | 2012-10-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Molecular Mechanism of Peroxisome Proliferator-Activated Receptor Alpha Activation by Wy14643: A New Mode of Ligand Recognition and Receptor Stabilization
J.Mol.Biol., 425, 2013
|
|
6HB7
 
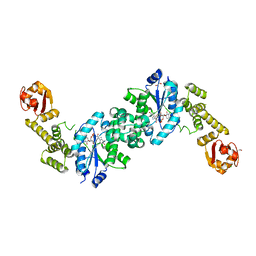 | | Crystal structure of E. coli tyrRS in complex with 5'-O-(N-L-tyrosyl)sulfamoyl-N3-methyluridine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine--tRNA ligase, ... | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
9GUF
 
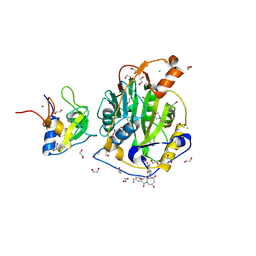 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571106 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
5TD5
 
 | | Crystal Structure of Human APOBEC3B variant complexed with ssDNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(P*TP*TP*CP*AP*T)-3'), ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MG2
 
 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
5K6R
 
 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a sulfonylamino-carbonyl-triazolinone herbicide, thiencarbazone-methyl | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2016-05-25 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2VIF
 
 | | Crystal structure of SOCS6 SH2 domain in complex with a c-KIT phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, MAST/STEM CELL GROWTH FACTOR RECEPTOR, SUPPRESSOR OF CYTOKINE SIGNALLING 6 | | Authors: | Bullock, A, Pike, A.C.W, Savitsky, P, Keates, T, Pilka, E.S, von Delft, F, Edwards, A, Weigelt, J, Arrowsmith, C.H, Knapp, S. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for C-Kit Inhibition by the Suppressor of Cytokine Signaling 6 (Socs6) Ubiquitin Ligase.
J.Biol.Chem., 286, 2011
|
|
6HDQ
 
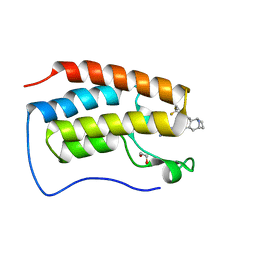 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH : 8-(((1R,2R,3R,5S)-2-(2-(4,4-difluorocyclohexyl)ethyl)-8-azabicyclo[3.2.1]octan-3-yl)amino)-3-methyl-5-(5-methylpyridin-3-yl)-1,7-naphthyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 8-(((1R,2R,3R,5S)-2-(2-(4,4-difluorocyclohexyl)ethyl)-8-azabicyclo[3.2.1]octan-3-yl)amino)-3-methyl-5-(5-methylpyridin-3-yl)-1,7-naphthyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2018-08-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Aiming to Miss a Moving Target: Bromo and Extra Terminal Domain (BET) Selectivity in Constrained ATAD2 Inhibitors.
J. Med. Chem., 61, 2018
|
|
5K9W
 
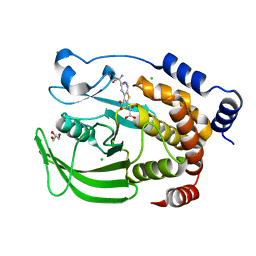 | | Protein Tyrosine Phosphatase 1B (1-301) in complex with TCS401, closed state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
6HE1
 
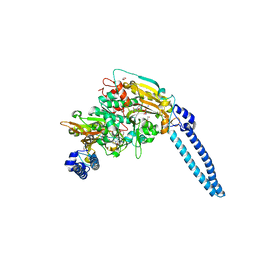 | | Pseudomonas aeruginosa Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine, SODIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
7B0D
 
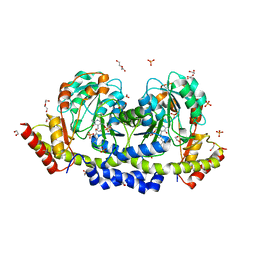 | | Sugar transaminase from Archaeoglobus veneficus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | James, P, Littlechild, J.A, De Rose, S.A, Isupov, M.N. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Sugar transaminases from hot environments
To Be Published
|
|
5CZZ
 
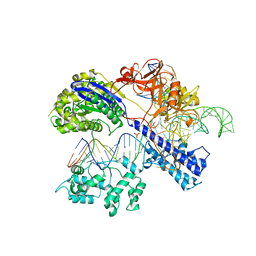 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGAAT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
4NYY
 
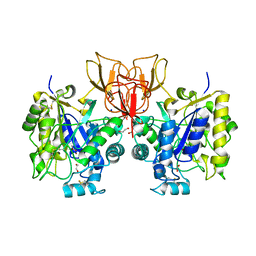 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 2 21 21 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5G5N
 
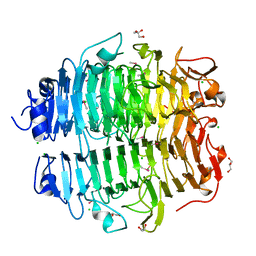 | | Structure of the snake adenovirus 1 hexon-interlacing LH3 protein, methylmercury chloride derivative | | Descriptor: | CHLORIDE ION, GLYCEROL, LH3 HEXON-INTERLACING CAPSID PROTEIN, ... | | Authors: | Nguyen, T.H, Singh, A.K, Albala-Perez, B, van Raaij, M.J. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid.
Structure, 25, 2017
|
|
5GI9
 
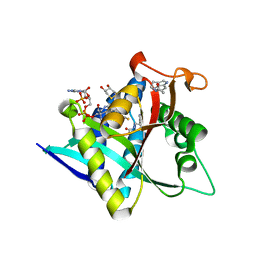 | | Crystal Structure of Drosophila melanogaster Dopamine N-Acetyltransferase in Ternary Complex with CoA and Acetyl-Tryptamine | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, COENZYME A, Dopamine N-acetyltransferase, ... | | Authors: | Yang, Y.C, Lin, S.J, Cheng, K.C, Cheng, H.C, Lyu, P.C. | | Deposit date: | 2016-06-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An essential role of acetyl coenzyme A in the catalytic cycle of insect arylalkylamine N-acetyltransferase.
Commun Biol, 3, 2020
|
|
5D2G
 
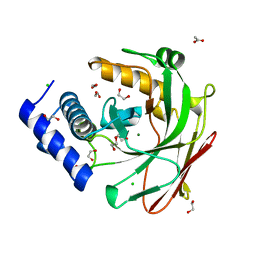 | |
6AGP
 
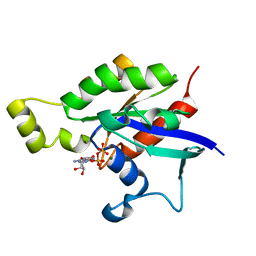 | | Structure of Rac1 in the low-affinity state for Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Toyama, Y, Kontani, K, Katada, T, Shimada, I. | | Deposit date: | 2018-08-13 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational landscape alternations promote oncogenic activities of Ras-related C3 botulinum toxin substrate 1 as revealed by NMR.
Sci Adv, 5, 2019
|
|
5D3X
 
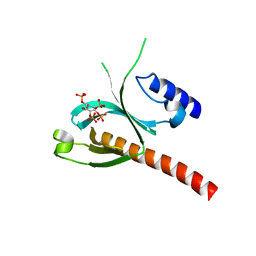 | | Crystal Structure of the P-Rex1 PH domain with Inositol-(1,3,4,5)-Tetrakisphosphate Bound | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
6AL8
 
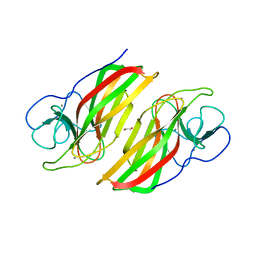 | | Crystal structure HpiC1 Y101F/F138S | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6QA6
 
 | | Glycogen Phosphorylase b in complex with 30 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-2-naphthalen-2-yl-8,9,10-tris(oxidanyl)-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
