8ZKK
 
 | |
6BW4
 
 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
4JAA
 
 | | Factor inhibiting HIF-1 alpha in complex with consensus ankyrin repeat domain-(d)LEU peptide | | Descriptor: | CONSENSUS ANKYRIN REPEAT DOMAIN-(d)LEU, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Scotti, J.S, Ge, W, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of an oxygenase in complex with substrate
To be Published
|
|
1HJP
 
 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | Descriptor: | RUVA | | Authors: | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 1997-08-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
8P2K
 
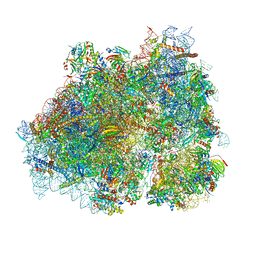 | | Ternary complex of translating ribosome, NAC and METAP1 | | Descriptor: | 18s rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jia, M, Jaskolowski, M, Scaiola, A, Jomaa, A, Ban, N. | | Deposit date: | 2023-05-16 | | Release date: | 2023-07-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | NAC controls cotranslational N-terminal methionine excision in eukaryotes.
Science, 380, 2023
|
|
5APW
 
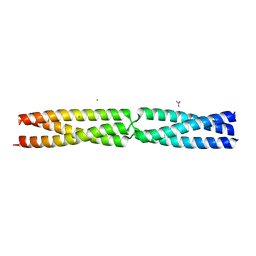 | | Sequence MATKDD inserted between GCN4 adaptors - Structure T6 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, ... | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
6GBM
 
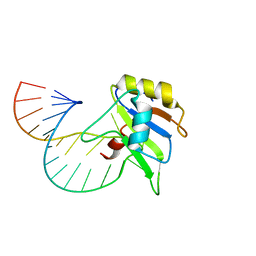 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
8BTK
 
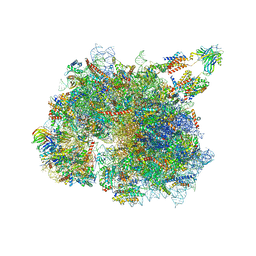 | | Structure of the TRAP complex with the Sec translocon and a translating ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Jaskolowski, M, Jomaa, A, Gamerdinger, M, Shrestha, S, Leibundgut, M, Deuerling, E, Ban, N. | | Deposit date: | 2022-11-29 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of the TRAP complex function in ER protein biogenesis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z8R
 
 | | CAND1-CUL1-RBX1 | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8V
 
 | | CAND1-SCF-SKP2 (SKP1deldel) CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8T
 
 | | CAND1-SCF-SKP2 CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
4REW
 
 | | Crystal structure of the non-phosphorylated human alpha1 beta2 gamma1 holo-AMPK complex | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Zhou, X.E, Ke, J, Li, X, Wang, L, Gu, X, de Waal, P.W, Tan, M.H.E, Wang, D, Wu, D, Xu, H.E, Melcher, K. | | Deposit date: | 2014-09-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.58 Å) | | Cite: | Structural basis of AMPK regulation by adenine nucleotides and glycogen.
Cell Res., 25, 2015
|
|
5APV
 
 | |
5APY
 
 | |
7DOA
 
 | |
1MY5
 
 | | NF-kappaB p65 subunit dimerization domain homodimer | | Descriptor: | NF-kappaB p65 (RelA) subunit | | Authors: | Huxford, T, Mishler, D, Phelps, C.B, Huang, D.-B, Sengchanthalangsy, L.L, Reeves, R, Hughes, C.A, Komives, E.A, Ghosh, G. | | Deposit date: | 2002-10-03 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Solvent exposed non-contacting amino acids play a critical role in NF-kappaB/I kappaB alpha complex formation
J.Mol.Biol., 324, 2002
|
|
4QFS
 
 | | Structure of AMPK in complex with Br2-A769662core activator and STAUROSPORINE inhibitor | | Descriptor: | 2-bromo-3-(4-bromophenyl)-4-hydroxy-6-oxo-6,7-dihydrothieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural Basis for AMPK Activation: Natural and Synthetic Ligands Regulate Kinase Activity from Opposite Poles by Different Molecular Mechanisms.
Structure, 22, 2014
|
|
6G99
 
 | | Solution structure of FUS-ZnF bound to UGGUG | | Descriptor: | RNA (5'-R(*UP*GP*GP*UP*G)-3'), RNA-binding protein FUS, ZINC ION | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
1EBO
 
 | | CRYSTAL STRUCTURE OF THE EBOLA VIRUS MEMBRANE-FUSION SUBUNIT, GP2, FROM THE ENVELOPE GLYCOPROTEIN ECTODOMAIN | | Descriptor: | CHLORIDE ION, EBOLA VIRUS ENVELOPE PROTEIN CHIMERA CONSISTING OF A FRAGMENT OF GCN4 ZIPPER CLONED N-TERMINAL TO A FRAGMENT OF GP2, ZINC ION | | Authors: | Weissenhorn, W, Carfi, A, Lee, K.H, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1998-11-03 | | Release date: | 1999-07-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain.
Mol.Cell, 2, 1998
|
|
7KXS
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5APX
 
 | | Sequence MATKDDIAN inserted between GCN4 adaptors - Structure T9(6) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PHOSPHATE ION | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
4HA8
 
 | | Structure of Staphylococcus aureus biotin protein ligase in complex with biotin acetylene | | Descriptor: | (3aS,4S,6aR)-4-(hex-5-yn-1-yl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Yap, M, Wilce, M, Polyak, S, Soares da Costa, T, Tieu, W. | | Deposit date: | 2012-09-25 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Staphylococcus aureus biotin protein ligase in complex with biotin acetylene
To be Published
|
|
2XHC
 
 | |
3NBH
 
 | | Crystal structure of human RMI1C-RMI2 complex | | Descriptor: | RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | Authors: | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
3PUP
 
 | | Structure of Glycogen Synthase Kinase 3 beta (GSK3B) in complex with a ruthenium octasporine ligand (OS1) | | Descriptor: | Glycogen synthase kinase-3 beta, Ruthenium octasporine | | Authors: | Filippakopoulos, P, Kraling, K, Essen, L.O, Meggers, E, Knapp, S. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structurally sophisticated octahedral metal complexes as highly selective protein kinase inhibitors.
J.Am.Chem.Soc., 133, 2011
|
|
