7L1Z
 
 | | Unlocking the structural features for the exo-xylobiosidase activity of an unusual GH11 member identified in a compost-derived consortium - NT-truncated form | | Descriptor: | 1,2-ETHANEDIOL, Exo-B-1,4-beta-xylanase, SULFATE ION | | Authors: | Kadowaki, M.A.S, Polikarpov, I, Briganti, L, Evangelista, D.E. | | Deposit date: | 2020-12-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unlocking the structural features for the xylobiohydrolase activity of an unusual GH11 member identified in a compost-derived consortium.
Biotechnol.Bioeng., 118, 2021
|
|
3M47
 
 | | Crystal structure of the mutant I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
1VYR
 
 | | Structure of pentaerythritol tetranitrate reductase complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-05 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
1FHW
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
4L5E
 
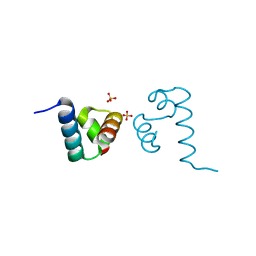 | | Crystal structure of A. aeolicus NtrC1 DNA binding domain | | Descriptor: | SULFATE ION, Transcriptional regulator (NtrC family) | | Authors: | Young, A, Maris, A.E, Vidangos, N.K, Hong, E, Pelton, J.G, Batchelor, J.D, Wemmer, D.E. | | Deposit date: | 2013-06-10 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure, function, and tethering of DNA-binding domains in sigma (54) transcriptional activators.
Biopolymers, 99, 2013
|
|
2O4V
 
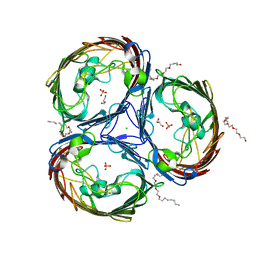 | | An arginine ladder in OprP mediates phosphate specific transfer across the outer membrane | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, T.F, Bains, M, Hancock, R.E, Strynadka, N.C. | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | An arginine ladder in OprP mediates phosphate-specific transfer across the outer membrane.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2O53
 
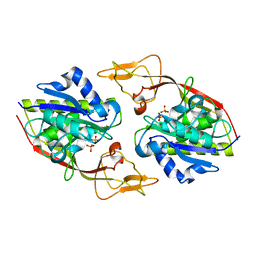 | | Crystal structure of apo-Aspartoacylase from human brain | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Le Coq, J, Pavlovsky, A, Sanishvili, R, Viola, R.E. | | Deposit date: | 2006-12-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Examination of the Mechanism of Human Brain Aspartoacylase through the Binding of an Intermediate Analogue.
Biochemistry, 47, 2008
|
|
1FN4
 
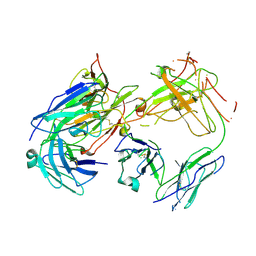 | | CRYSTAL STRUCTURE OF FAB198, AN EFFICIENT PROTECTOR OF ACETYLCHOLINE RECEPTOR AGAINST MYASTHENOGENIC ANTIBODIES | | Descriptor: | MONOCLONAL ANTIBODY AGAINST ACETYLCHOLINE RECEPTOR | | Authors: | Poulas, K, Eliopoulos, E, Vatzaki, E, Navaza, J, Kontou, M. | | Deposit date: | 2000-08-21 | | Release date: | 2001-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Fab198, an efficient protector of the acetylcholine receptor against myasthenogenic antibodies.
Eur.J.Biochem., 268, 2001
|
|
2ITE
 
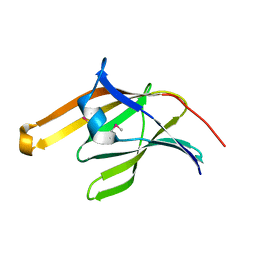 | | Crystal structure of the IsdA NEAT domain from Staphylococcus aureus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Iron-regulated surface determinant protein A | | Authors: | Grigg, J.C, Vermeiren, C.L, Heinrichs, D.E, Murphy, M.E. | | Deposit date: | 2006-10-19 | | Release date: | 2006-12-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Haem recognition by a Staphylococcus aureus NEAT domain.
Mol.Microbiol., 63, 2007
|
|
2NX4
 
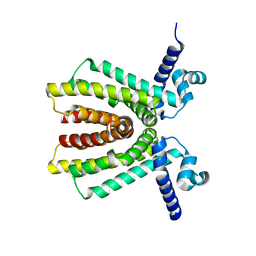 | | The Crystal Structure of athe Putative TetR-family transcriptional regulator Rha06780 from Rhodococcus sp. Rha1. | | Descriptor: | Transcriptional regulator, TetR family protein | | Authors: | Zhang, R, Evdokimova, E, Kudritskam, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of a TetR-family transcriptional regulator from Rhodococcus sp.
To be Published, 2006
|
|
4YH8
 
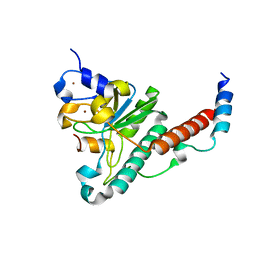 | | Structure of yeast U2AF complex | | Descriptor: | Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ZINC ION | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2015-02-27 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel 3' splice site recognition by the two zinc fingers in the U2AF small subunit.
Genes Dev., 29, 2015
|
|
1BIX
 
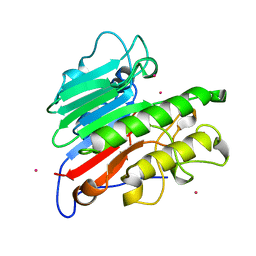 | | THE CRYSTAL STRUCTURE OF THE HUMAN DNA REPAIR ENDONUCLEASE HAP1 SUGGESTS THE RECOGNITION OF EXTRA-HELICAL DEOXYRIBOSE AT DNA ABASIC SITES | | Descriptor: | AP ENDONUCLEASE 1, PLATINUM (II) ION, SAMARIUM (III) ION | | Authors: | Gorman, M.A, Morera, S, Rothwell, D.G, De La Fortelle, E, Mol, C.D, Tainer, J.A, Hickson, I.D, Freemont, P.S. | | Deposit date: | 1998-06-19 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites.
EMBO J., 16, 1997
|
|
3HMI
 
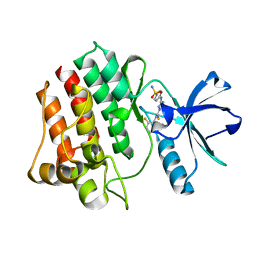 | | The crystal structure of human ABL2 in complex with 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Tyrosine-protein kinase ABL2 | | Authors: | Ugochukwu, E, Salah, E, Barr, A, Mahajan, P, Shrestha, B, Savitsky, P, Chaikuad, A, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of human ABL2 in complex with 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE
To be Published
|
|
2C8C
 
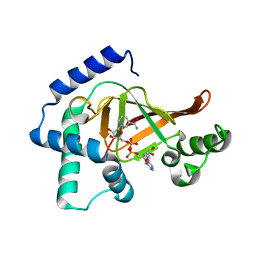 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (NAD-bound state, crystal form I) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2VKD
 
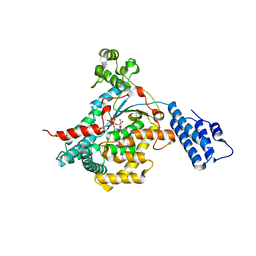 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP-GLC AND MANGANESE ION | | Descriptor: | CYTOTOXIN L, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ziegler, M.O.P, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Conformational Changes and Reaction of Clostridial Glycosylating Toxins.
J.Mol.Biol., 377, 2008
|
|
2V7A
 
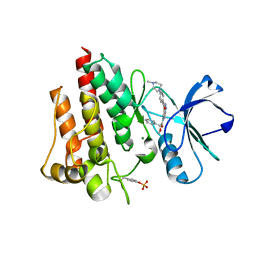 | | Crystal structure of the T315I Abl mutant in complex with the inhibitor PHA-739358 | | Descriptor: | MAGNESIUM ION, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Modugno, M, Casale, E, Soncini, C, Rosettani, P, Colombo, R, Lupi, R, Rusconi, L, Fancelli, D, Carpinelli, P, Cameron, A.D, Isacchi, A, Moll, J. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the T315I Abl Mutant in Complex with the Aurora Kinases Inhibitor Pha-739358.
Cancer Res., 67, 2007
|
|
5E40
 
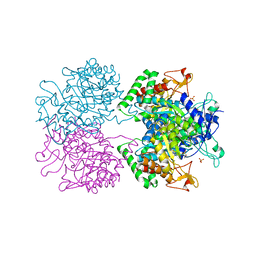 | | 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis with D-tyrosine bound in the phenylalanine binding site | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, D-TYROSINE, GLYCEROL, ... | | Authors: | Reichau, S, Jiao, W, Parker, E.J. | | Deposit date: | 2015-10-05 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the Sophisticated Synergistic Allosteric Regulation of Aromatic Amino Acid Biosynthesis in Mycobacterium tuberculosis Using -Amino Acids.
Plos One, 11, 2016
|
|
6A77
 
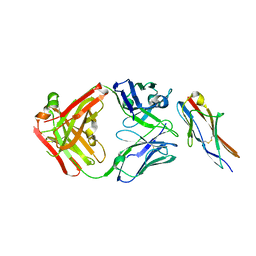 | | Crystal structure of the fifth immunoglobulin domain (Ig5) of human Robo1 in complex with the Fab fragment of murine monoclonal antibody B5209B | | Descriptor: | Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, Roundabout homolog 1 | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2019-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
4CSB
 
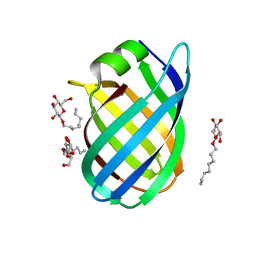 | | Structure of the Virulence-Associated Protein VapD from the intracellular pathogen Rhodococcus equi. | | Descriptor: | VIRULENCE ASSOCIATED PROTEIN VAPD, octyl beta-D-glucopyranoside | | Authors: | Whittingham, J.L, Blagova, E.V, Finn, C.E, Luo, H, Miranda-CasoLuengo, R, Turkenburg, J.P, Leech, A.P, Walton, P.H, Meijers, W.G, Wilkinson, A.J. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Virulence-Associated Protein Vapd from the Intracellular Pathogen Rhodococcus Equi.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1HSE
 
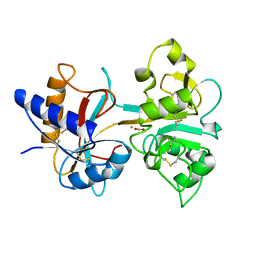 | | H253M N TERMINAL LOBE OF HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Nicholson, H, Anderson, B.F, Baker, E.N. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutagenesis of the histidine ligand in human lactoferrin: iron binding properties and crystal structure of the histidine-253-->methionine mutant.
Biochemistry, 36, 1997
|
|
4YIQ
 
 | | Structure of the CEACAM6-CEACAM8 heterodimer | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 6, Carcinoembryonic antigen-related cell adhesion molecule 8, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Diverse oligomeric states of CEACAM IgV domains.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2OFE
 
 | | The Crystal structure of Sclerotium rolfsii lectin in complex with N-acetyl-D-glucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Carbohydrate Recognition of the Sclerotium rolfsii Lectin
J.Mol.Biol., 368, 2007
|
|
1WB8
 
 | | Iron Superoxide Dismutase (FE-SOD) from the Hyperthermophile SULFOLOBUS SOLFATARICUS. 2.3 A Resolution Structure of Recombinant Protein with a Covalently Modified Tyrosine in the Active Site. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE [FE], phenylmethanesulfonic acid | | Authors: | Ursby, T, Adinolfi, B.S, Al-Karadaghi, S, De Vendittis, E, Bocchini, V. | | Deposit date: | 2004-10-31 | | Release date: | 2004-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Iron Superoxide Dismutase from the Archaeon Sulfolobus Solfataricus: Analysis of Structure and Thermostability
J.Mol.Biol., 286, 1999
|
|
1BL6
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
3MW7
 
 | |
