7MFZ
 
 | |
6ZR8
 
 | |
8DNZ
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by apratoxin F | | Descriptor: | Apratoxin F peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
6V4E
 
 | | Crystal Structure Analysis of Zebra Fish MDM | | Descriptor: | Protein Mdm4, Stapled peptide QSQQTF(0EH)NLWRLL(MK8)QN(NH2) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-27 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Structural Determinant for Selective Targeting of HDMX.
Structure, 28, 2020
|
|
6ME8
 
 | | XFEL crystal structure of human melatonin receptor MT2 (N86D) in complex with 2-phenylmelatonin | | Descriptor: | N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
7MQY
 
 | | Zebrafish CNTN4 APLP2 complex | | Descriptor: | Fusion protein of CNTN4 and APLP2 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
6V5C
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
7MRQ
 
 | |
6UQJ
 
 | |
6P1P
 
 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and bound incoming dCMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
8DOC
 
 | | Crystal structure of RPE65 in complex with compound 16e and palmitate | | Descriptor: | (1R)-1-[3-(cyclohexylmethoxy)phenyl]-3-(methylamino)propan-1-ol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FE (II) ION, ... | | Authors: | Bassetto, M, Kiser, P.D. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tuning the Metabolic Stability of Visual Cycle Modulators through Modification of an RPE65 Recognition Motif.
J.Med.Chem., 66, 2023
|
|
6MEI
 
 | |
7MRO
 
 | | Zebrafish CNTN4 FN1-FN3 domains | | Descriptor: | Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-07 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
6P1V
 
 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing undamaged template dG and bound incoming dCMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
8EA6
 
 | | NKG2D complexed with inhibitor 3e | | Descriptor: | N-{(1S)-2-(dimethylamino)-2-oxo-1-[3-(trifluoromethyl)phenyl]ethyl}-4'-(trifluoromethyl)[1,1'-biphenyl]-2-carboxamide, NKG2-D type II integral membrane protein | | Authors: | Thompson, A.A, Grant, J.C, Karpowich, N.K, Sharma, S. | | Deposit date: | 2022-08-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of small-molecule protein-protein interaction inhibitors for NKG2D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7A9C
 
 | |
6UQN
 
 | |
7MRS
 
 | | Zebrafish CNTN4 APPb complex | | Descriptor: | Amyloid-beta A4 protein, Contactin-4 | | Authors: | Bouyain, S, Karuppan, S.J. | | Deposit date: | 2021-05-09 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Members of the vertebrate contactin and amyloid precursor protein families interact through a conserved interface.
J.Biol.Chem., 298, 2021
|
|
6UQS
 
 | |
8DNX
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by cotransin | | Descriptor: | Cotransin analogue peptide inhibitor, Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
6MFM
 
 | |
7AA9
 
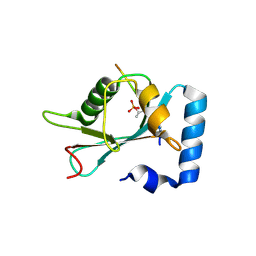 | | Structure of SCOC pT13/pT15 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pT13/PT15 SCOC LIR | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
6P24
 
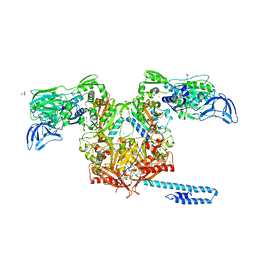 | | Escherichia coli tRNA synthetase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Kahne, D, Baidin, V, Owens, T.W. | | Deposit date: | 2019-05-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Simple Secondary Amines Inhibit Growth of Gram-Negative Bacteria through Highly Selective Binding to Phenylalanyl-tRNA Synthetase.
J.Am.Chem.Soc., 143, 2021
|
|
6MEN
 
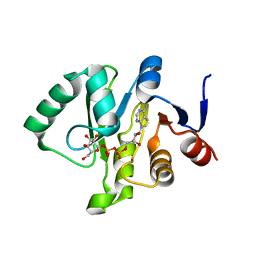 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6V6G
 
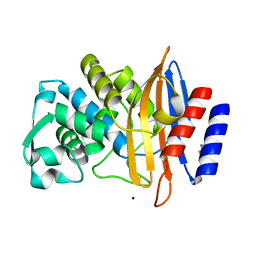 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
