7OI0
 
 | | E.coli delta rbfA pre-30S ribosomal subunit class D | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Maksimova, E, Korepanov, A, Baymukhametov, T, Kravchenko, O, Stolboushkina, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center.
Int J Mol Sci, 22, 2021
|
|
5APZ
 
 | | Thermosinus carboxydivorans Nor1 Tcar0761 residues 68-101 and 191-211 fused to GCN4 adaptors | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, NOR1 TCAR0761 | | Authors: | Hartmann, M.D, Deiss, S, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
8EFJ
 
 | |
7DOB
 
 | |
7DOA
 
 | |
1IJ1
 
 | |
2YNF
 
 | | HIV-1 Reverse Transcriptase Y188L mutant in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
5APX
 
 | | Sequence MATKDDIAN inserted between GCN4 adaptors - Structure T9(6) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PHOSPHATE ION | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APW
 
 | | Sequence MATKDD inserted between GCN4 adaptors - Structure T6 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, ... | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
2XIY
 
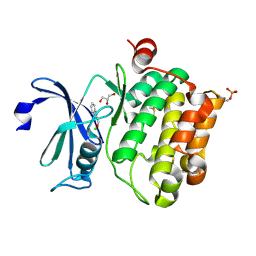 | | Protein kinase Pim-1 in complex with fragment-2 from crystallographic fragment screen | | Descriptor: | 2-HYDROXYMETHYL-BENZOIMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5E57
 
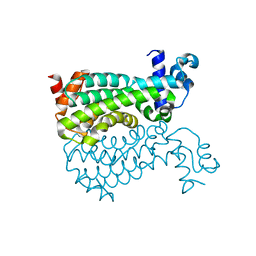 | | Crystal structure of Mycobacterium smegmatis AmtR | | Descriptor: | ACETATE ION, Transcription regulator AmtR | | Authors: | Vickers, C.J, McKenzie, J.L, Arcus, V.L. | | Deposit date: | 2015-10-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Function of AmtR in Mycobacterium smegmatis: Implications for Post-Transcriptional Regulation of Urea Metabolism through a Small Antisense RNA.
J.Mol.Biol., 428, 2016
|
|
5APV
 
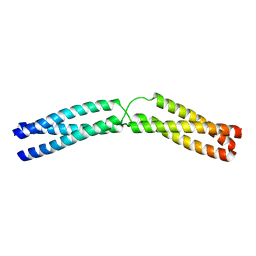 | |
5APY
 
 | |
2YNI
 
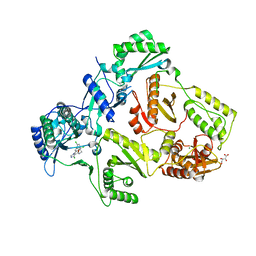 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK952 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-15 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2YNG
 
 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, MAGNESIUM ION, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
6N7G
 
 | | Cryo-EM structure of tetrameric Ptch1 in complex with ShhNp (form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
6N7K
 
 | | Cryo-EM structure of tetrameric Ptch1 in complex with ShhNp (form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
2YNH
 
 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK500 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-2-(hydroxymethyl)-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
5Q0O
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[4-(benzenecarbonyl)phenyl]-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, CHLORIDE ION, ... | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q14
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}-3-fluorobenzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2XJ2
 
 | | Protein kinase Pim-1 in complex with small molecule inhibitor | | Descriptor: | (2E)-3-{3-[6-(4-methyl-1,4-diazepan-1-yl)pyrazin-2-yl]phenyl}prop-2-enoic acid, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5Q0U
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1C
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2XJ0
 
 | | Protein kinase Pim-1 in complex with fragment-4 from crystallographic fragment screen | | Descriptor: | (E)-3-(2-AMINO-PYRIDINE-5YL)-ACRYLIC ACID, PROTO-ONCOGENE SERINE/THREONINE PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XJ1
 
 | | Protein kinase Pim-1 in complex with small molecule inibitor | | Descriptor: | (2E)-3-(3-{6-[(TRANS-4-AMINOCYCLOHEXYL)AMINO]PYRAZIN-2-YL}PHENYL)PROP-2-ENOIC ACID, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Schulz, M.N, Fanghanel, J, Schafer, M, Badock, V, Briem, H, Boemer, U, Nguyen, D, Husemann, M, Hillig, R.C. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystallographic Fragment Screen Identifies Cinnamic Acid Derivatives as Starting Points for Potent Pim-1 Inhibitors
Acta Crystallogr.,Sect.D, 67, 2011
|
|
