6TU7
 
 | | Structure of PfMyoA decorated Plasmodium Act1 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-1, Jasplakinolide, ... | | Authors: | Vahokoski, J, Calder, L.J, Lopez, A.J, Rosenthal, P.B, Kursula, I. | | Deposit date: | 2020-01-03 | | Release date: | 2021-01-13 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution structures of malaria parasite actomyosin and actin filaments.
Plos Pathog., 18, 2022
|
|
2I7O
 
 | | Structure of Re(4,7-dimethyl-phen)(Thr124His)(Lys122Trp)(His83Gln)AzCu(II), a Rhenium modified Azurin mutant | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Sudhamsu, J, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tryptophan-accelerated electron flow through proteins.
Science, 320, 2008
|
|
2HVX
 
 | | Discovery of Potent, Orally Active, Nonpeptide Inhibitors of Human Mast Cell Chymase by Using Structure-Based Drug Design | | Descriptor: | Chymase, [(1S)-1-(5-CHLORO-1-BENZOTHIEN-3-YL)-2-(2-NAPHTHYLAMINO)-2-OXOETHYL]PHOSPHONIC ACID | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond, H.R, de Garavilla, L, Wang, Y, Minor, L.A, Wells, G.I, Di Cera, E, Cantwell, A.M, Savvides, S.N, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of potent, selective, orally active, nonpeptide inhibitors of human mast cell chymase.
J.Med.Chem., 50, 2007
|
|
2I3W
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of S729C mutant | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT 2, GLUTAMIC ACID | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
6M65
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPNP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2I4B
 
 | | Crystal structure of Bicarbonate Transport Protein CmpA from Synechocystis sp. PCC 6803 in complex with bicarbonate and calcium | | Descriptor: | BICARBONATE ION, Bicarbonate transporter, CALCIUM ION | | Authors: | Koropatkin, N.M, Smith, T.J, Pakrasi, H.B. | | Deposit date: | 2006-08-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Structure of a Cyanobacterial Bicarbonate Transport Protein, CmpA.
J.Biol.Chem., 282, 2007
|
|
6LKE
 
 | | in meso full-length rat KMO in complex with an inhibitor identified via DNA-encoded chemical library screening | | Descriptor: | 4-chloranyl-2-[[5-chloranyl-2-(5-methoxy-1,3-dihydroisoindol-2-yl)-1,3-thiazol-4-yl]carbonyl-methyl-amino]-5-fluoranyl-benzoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mimasu, S, Yamagishi, H, Kiyohara, M, Hupp, D.C, Liu, J, Kakefuda, K, Okuda, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition.
Commun Biol, 4, 2021
|
|
6NLG
 
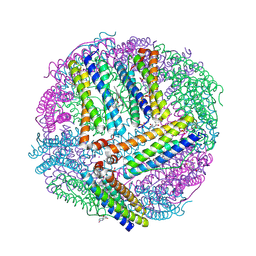 | | 1.50 A resolution structure of BfrB (C89S/K96C) from Pseudomonas aeruginosa in complex with a small molecule fragment (analog 1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-hydroxy-1H-isoindole-1,3(2H)-dione, Bacterioferritin, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6NU6
 
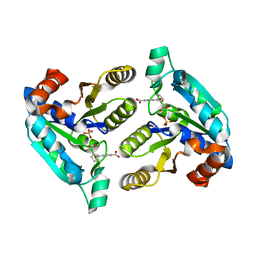 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 5 | | Descriptor: | 4-[(1R,2S)-2-(carboxymethyl)cyclopentane-1-carbonyl]benzoic acid, 4-[(1S,2R)-2-(carboxymethyl)cyclopentane-1-carbonyl]benzoic acid, ATP-dependent dethiobiotin synthetase BioD, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-01-31 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 5
To Be Published
|
|
2I2B
 
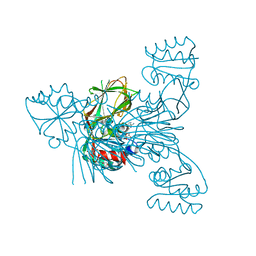 | |
2I4H
 
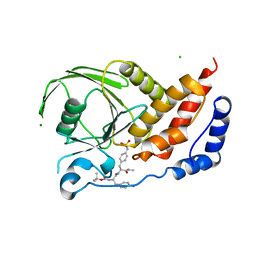 | | Structural studies of protein tyrosine phosphatase beta catalytic domain co-crystallized with a sulfamic acid inhibitor | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-(TERT-BUTOXYCARBONYL)-L-TYROSYL-N-METHYL-4-(SULFOAMINO)-L-PHENYLALANINAMIDE, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2HZL
 
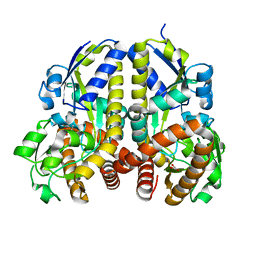 | | Crystal structures of a sodium-alpha-keto acid binding subunit from a TRAP transporter in its closed forms | | Descriptor: | PYRUVIC ACID, SODIUM ION, TRAP-T family sorbitol/mannitol transporter, ... | | Authors: | Gonin, S, Arnoux, P, Pierru, B, Alonso, B, Sabaty, M, Pignol, D. | | Deposit date: | 2006-08-09 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding.
Bmc Struct.Biol., 7, 2007
|
|
6O1J
 
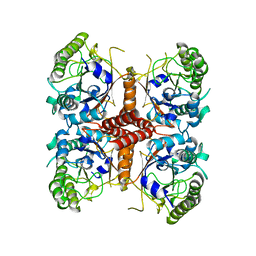 | |
6O1A
 
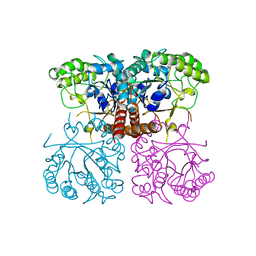 | |
2I49
 
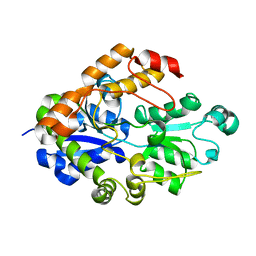 | |
2IBG
 
 | |
2I88
 
 | |
2FRX
 
 | | Crystal structure of YebU, a m5C RNA methyltransferase from E.coli | | Descriptor: | Hypothetical protein yebU | | Authors: | Erlandsen, H, Nordlund, P, Hallberg, B.M, Johnson, K.A, Ericsson, U.B. | | Deposit date: | 2006-01-20 | | Release date: | 2006-08-29 | | Last modified: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain
J.Mol.Biol., 360, 2006
|
|
2B1P
 
 | | inhibitor complex of JNK3 | | Descriptor: | 3-{6-[(2-CHLOROPHENYL)AMINO]-1H-INDAZOL-3-YL}-5-{[4-(DIMETHYLAMINO)BUTANOYL]AMINO}BENZOIC ACID, BETA-MERCAPTOETHANOL, Mitogen-activated protein kinase 10, ... | | Authors: | Swahn, B.M, Huerta, F, Kallin, E, Malmstrom, J, Weigelt, T, Viklund, J, Womack, P, Xue, Y, Ohberg, L. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of 6-anilinoindazoles as selective inhibitors of c-Jun N-terminal kinase-3
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2FPY
 
 | | Dual binding mode of a novel series of DHODH inhibitors | | Descriptor: | 3-({[3,5-DIFLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)THIOPHENE-2-CARBOXYLIC ACID, ACETATE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Baumgartner, R, Leban, J. | | Deposit date: | 2006-01-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J.Med.Chem., 49, 2006
|
|
6MNM
 
 | | 6256 TCR bound to I-Ab Padi4 | | Descriptor: | 6256 TCR alpha chain, 6256 TCR beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Blevins, S.J, Stadinski, B.D, Huseby, E.S. | | Deposit date: | 2018-10-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A temporal thymic selection switch and ligand binding kinetics constrain neonatal Foxp3+Tregcell development.
Nat.Immunol., 20, 2019
|
|
2G3J
 
 | | Structure of S.olivaceoviridis xylanase Q88A/R275A mutant | | Descriptor: | PHOSPHATE ION, Xylanase, alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Diertavitian, S, Kaneko, S, Fujimoto, Z, Kuno, A, Johansson, E, Lo Leggio, L. | | Deposit date: | 2006-02-20 | | Release date: | 2007-03-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based engineering of glucose specificity in a family 10 xylanase from Streptomyces olivaceoviridis E-86
PROCESS BIOCHEM, 47, 2012
|
|
2FQI
 
 | | dual binding modes of a novel series of DHODH inhibitors | | Descriptor: | 2-({[2,3,5,6-TETRAFLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENTA-1,3-DIENE-1-CARBOXYLIC ACID, ACETATE ION, Dihydroorotate dehydrogenase, ... | | Authors: | Baumgartner, R, Leban, J. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J.Med.Chem., 49, 2006
|
|
6N0U
 
 | |
6MT6
 
 | |
