2EKP
 
 | |
1QCA
 
 | |
4N9A
 
 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N97
 
 | | E. coli sliding clamp in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
8W87
 
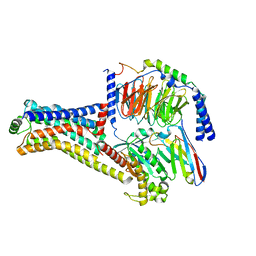 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
2FTY
 
 | |
2FVK
 
 | |
4K74
 
 | |
1AZW
 
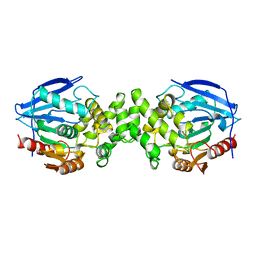 | | PROLINE IMINOPEPTIDASE FROM XANTHOMONAS CAMPESTRIS PV. CITRI | | Descriptor: | PROLINE IMINOPEPTIDASE | | Authors: | Medrano, F.J, Alonso, J, Garcia, J.L, Romero, A, Bode, W, Gomis-Ruth, F.X. | | Deposit date: | 1997-11-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of proline iminopeptidase from Xanthomonas campestris pv. citri: a prototype for the prolyl oligopeptidase family.
EMBO J., 17, 1998
|
|
4N99
 
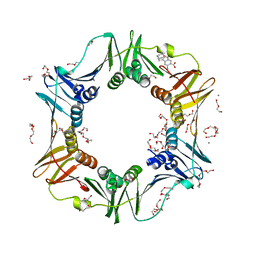 | | E. coli sliding clamp in complex with 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid | | Descriptor: | 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
2FVM
 
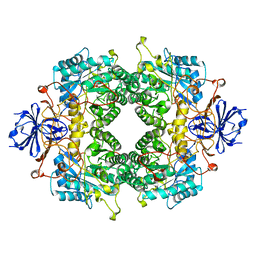 | |
8FDL
 
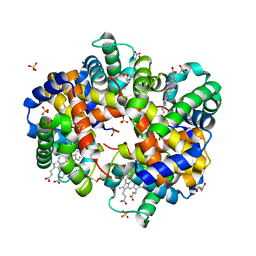 | | Human Hemoglobin with Nitrosochloramphenicol | | Descriptor: | GLYCEROL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Powell, S.M, Richter-Addo, G.B, Thomas, L.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structural investigations of heme protein derivatives resulting from reactions of aryl- and alkylhydroxylamines with human hemoglobin.
J.Inorg.Biochem., 246, 2023
|
|
4N94
 
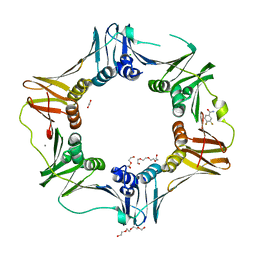 | | E. coli sliding clamp in complex with 3,4-difluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 3,4-difluorobenzamide, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
6CGL
 
 | |
4C9K
 
 | |
4N98
 
 | |
4C9N
 
 | |
1SXJ
 
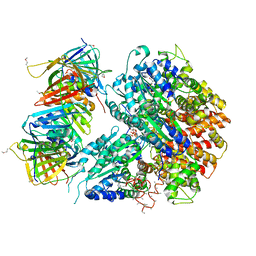 | | Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Activator 1 37 kDa subunit, Activator 1 40 kDa subunit, ... | | Authors: | Bowman, G.D, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex.
Nature, 429, 2004
|
|
2RCX
 
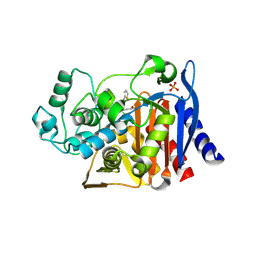 | | AmpC Beta-lactamase in complex with (1R)-1-(2-Thiophen-2-yl-acetylamino)-1-(3-(2-carboxyvinyl)-phenyl) methylboronic acid | | Descriptor: | (1R)-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-1-(3-(2-CARBOXYVINYL)-PHENYL) METHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION | | Authors: | Morandi, F, Morandi, S, Prati, F, Shoichet, B.K. | | Deposit date: | 2007-09-20 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based optimization of cephalothin-analogue boronic acids as beta-lactamase inhibitors
Bioorg.Med.Chem., 16, 2008
|
|
1OK7
 
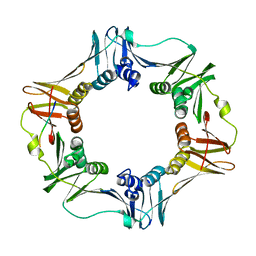 | | A Conserved protein binding-site on Bacterial Sliding Clamps | | Descriptor: | DNA POLYMERASE III, DNA POLYMERASE IV | | Authors: | Burnouf, D.Y, Olieric, V, Wagner, J, Fujii, S, Reinbolt, J, Fuchs, R.P.P, Dumas, P. | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of Sliding Clamp/Ligand Interactions Suggest a Competition between Replicative and Translesion DNA Polymerases
J.Mol.Biol., 335, 2004
|
|
2EKQ
 
 | |
3H03
 
 | |
1MMN
 
 | | X-RAY STRUCTURES OF THE MGADP, MGATPGAMMAS, AND MGAMPPNP COMPLEXES OF THE DICTYOSTELIUM DISCOIDEUM MYOSIN MOTOR DOMAIN | | Descriptor: | MAGNESIUM ION, MYOSIN, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Rayment, I. | | Deposit date: | 1997-07-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain.
Biochemistry, 36, 1997
|
|
3ZH9
 
 | | Bacillus subtilis DNA clamp loader delta protein (YqeN) | | Descriptor: | DELTA, GLYCEROL, SULFATE ION | | Authors: | Suwannachart, C, Sedelnikova, S, Soultanas, P, Oldham, N.J, Rafferty, J.B. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structure and Assembly of the Bacillus Subtilis Clamp-Loader Complex and its Interaction with the Replicative Helicase.
Nucleic Acids Res., 41, 2013
|
|
2PKE
 
 | |
