6DER
 
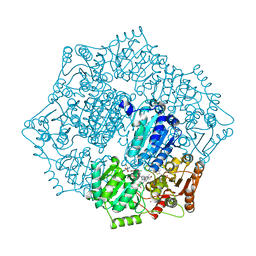 | | Crystal structure of Candida albicans acetohydroxyacid synthase in complex with the herbicide metosulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Guddat, L.W. | | Deposit date: | 2018-05-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.126 Å) | | Cite: | Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ZZY
 
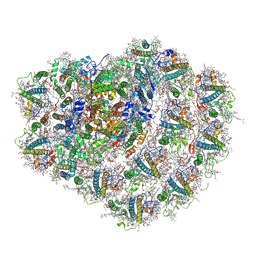 | | Structure of high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
2IHU
 
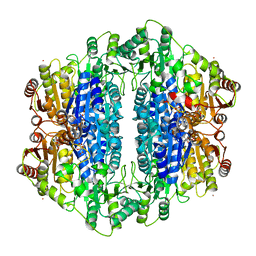 | | Carboxyethylarginine synthase from Streptomyces clavuligerus: putative reaction intermediate complex | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-2-[(1Z)-1-HYDROXY-3-(PHOSPHONOOXY)PROP-1-EN-1-YL]-3-{[(4Z)-4-IMINO-2- METHYL-4,5-DIHYDROPYRIMIDIN-5-YL]METHYL}-4-METHYL-1,3-THIAZOL-3-IUM, Carboxyethylarginine synthase, ... | | Authors: | Caines, M.E, Schofield, C.J. | | Deposit date: | 2006-09-27 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mechanistic studies on N(2)-(2-carboxyethyl)arginine synthase.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
5FSC
 
 | | Structure of tectonin 2 from Laccaria bicolor in complex with allyl- alpha_4-methyl-mannoside | | Descriptor: | MAGNESIUM ION, TECTONIN 2, prop-2-en-1-yl 4-O-methyl-alpha-D-mannopyranoside | | Authors: | Sommer, R, Bleuer, S, Titz, A, Kunzler, M, Varrot, A. | | Deposit date: | 2016-01-04 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Fungal Tectonin in Complex with O-Methylated Glycans Suggest Key Role in Innate Immune Defense.
Structure, 26, 2018
|
|
1AGM
 
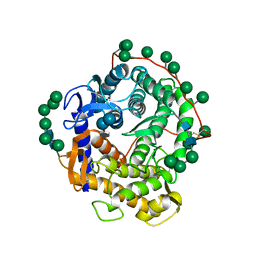 | | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-05-13 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. X100 to 2.4-A resolution.
J.Biol.Chem., 269, 1994
|
|
3POC
 
 | | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Tan, K, Tesar, C, Wilton, R, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-22 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
To be Published
|
|
1OSE
 
 | | Porcine pancreatic alpha-amylase complexed with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Gilles, C, Payan, F. | | Deposit date: | 1996-03-20 | | Release date: | 1997-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of pig pancreatic alpha-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose.
Eur.J.Biochem., 238, 1996
|
|
7U3D
 
 | | Structure of S. venezuelae GlgX-c-di-GMP-acarbose complex (4.6) | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Glycogen debranching enzyme GlgX | | Authors: | Schumacher, M.A. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric regulation of glycogen breakdown by the second messenger cyclic di-GMP.
Nat Commun, 13, 2022
|
|
7LID
 
 | |
7LHV
 
 | | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SULFATE ION, ... | | Authors: | Wang, L, Chen, K, Zhou, M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and function of an Arabidopsis thaliana sulfate transporter.
Nat Commun, 12, 2021
|
|
7LXA
 
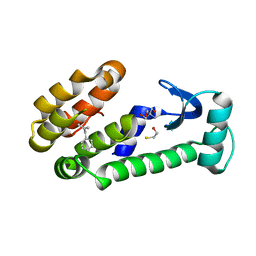 | | T4 lysozyme mutant L99A | | Descriptor: | (2-methylprop-2-en-1-yl)benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-03-03 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LSQ
 
 | |
2ZBH
 
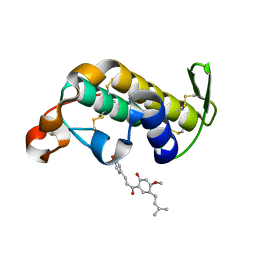 | | Crystal structure of the complex of phospholipase A2 with Bavachalcone from Aerva lanata at 2.6 A resolution | | Descriptor: | (2E)-1-[2-hydroxy-4-methoxy-5-(3-methylbut-2-en-1-yl)phenyl]-3-(4-hydroxyphenyl)prop-2-en-1-one, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Damodar, N.C, Jain, R, Singh, N, Sharma, S, Kaur, P, Haridas, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-10-20 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of phospholipase A2 with Bavachalcone from Aerva lanata at 2.6 A resolution
To be Published
|
|
3PHA
 
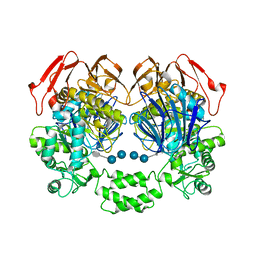 | | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose
To be Published
|
|
3SY0
 
 | | S25-2- A(2-8)-A(2-4)KDO trisaccharide complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S25-2 FAB (IGG1K) heavy chain, ... | | Authors: | Nguyen, H.P, Seto, N.O, Mackenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes
NAT.STRUCT.BIOL., 10, 2003
|
|
8WB4
 
 | | Structure of PSII-ACPII supercomplex from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-09-08 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
4M7J
 
 | | Crystal structure of S25-26 in complex with Kdo(2.8)Kdo(2.4)Kdo trisaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Haji-Ghassemi, O, Evans, S.V, Muller-Loennies, S, Saldova, R, Muniyappa, M, Brade, L, Rudd, P.M, Harvey, D.J, Kosma, P, Brade, H. | | Deposit date: | 2013-08-12 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Groove-type Recognition of Chlamydiaceae-specific Lipopolysaccharide Antigen by a Family of Antibodies Possessing an Unusual Variable Heavy Chain N-Linked Glycan.
J.Biol.Chem., 289, 2014
|
|
3BPC
 
 | | co-crystal structure of S25-2 Fab in complex with 5-deoxy-4-epi-2,3-dehydro Kdo (4.8) Kdo | | Descriptor: | 3,4,5-trideoxy-alpha-D-erythro-oct-3-en-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Fab, antibody fragment (IgG1k), ... | | Authors: | Brooks, C.L, Evans, S.V. | | Deposit date: | 2007-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes.
J.Mol.Biol., 377, 2008
|
|
3TOP
 
 | | Crystral Structure of the C-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltase-glucoamylase, intestinal | | Authors: | Shen, Y, Qin, X.H, Ren, L.M. | | Deposit date: | 2011-09-06 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Structural insight into substrate specificity of human intestinal maltase-glucoamylase
Protein Cell, 2, 2011
|
|
5VD5
 
 | |
5VDA
 
 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-101, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1S)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5VD9
 
 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-097, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1R)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5HQA
 
 | | A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glucosidase, CALCIUM ION, ... | | Authors: | Li, J, He, C, Xiao, Y. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
5VD8
 
 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-099, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-chloro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5X7Q
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
