7D5Y
 
 | |
3HO6
 
 | |
3PEE
 
 | |
6EN3
 
 | | Crystal structure of full length EndoS from Streptococcus pyogenes in complex with G2 oligosaccharide. | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2,Multifunctional-autoprocessing repeats-in-toxin, NICKEL (II) ION, ... | | Authors: | Trastoy, B, Klontz, E.H, Orwenyo, J, Marina, A, Wang, L.X, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural basis for the recognition of complex-type N-glycans by Endoglycosidase S.
Nat Commun, 9, 2018
|
|
3PA8
 
 | |
3EEB
 
 | | Structure of the V. cholerae RTX cysteine protease domain | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION | | Authors: | Lupardus, P.J, Shen, A, Bogyo, M, Garcia, K.C. | | Deposit date: | 2008-09-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain
Science, 322, 2008
|
|
3FZY
 
 | | Crystal Structure of Pre-cleavage Form of Cysteine Protease Domain from Vibrio cholerae RtxA Toxin | | Descriptor: | CHLORIDE ION, INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, ... | | Authors: | Shuvalova, L, Minasov, G, Prochazkova, K, Satchell, K.J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites
J.Biol.Chem., 284, 2009
|
|
3GCD
 
 | | Structure of the V. cholerae RTX cysteine protease domain in complex with an aza-Leucine peptide inhibitor | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, SODIUM ION, ... | | Authors: | Lupardus, P.J, Garcia, K.C, Shen, A, Bogyo, M. | | Deposit date: | 2009-02-21 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanistic and structural insights into the proteolytic activation of Vibrio cholerae MARTX toxin.
Nat.Chem.Biol., 5, 2009
|
|
8YJA
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain lacking beta-flap | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MARTX cysteine protease domain, SODIUM ION | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
9BJA
 
 | |
8YJC
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain C3727A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INOSITOL HEXAKISPHOSPHATE, Multifunctional autoprocessing repeat-in-toxin (MARTX), ... | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
4R04
 
 | |
7POG
 
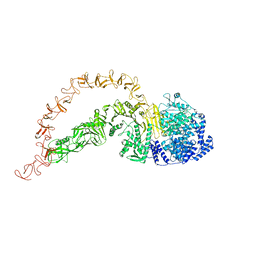 | | High-resolution structure of native toxin A from Clostridioides difficile | | Descriptor: | Toxin A, ZINC ION | | Authors: | Boesen, T, Joergensen, R, Aminzadeh, A, Engelbrecht Larsen, C. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structure of native toxin A from Clostridioides difficile.
Embo Rep., 23, 2022
|
|
7V1N
 
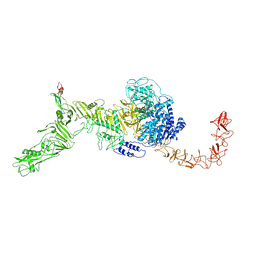 | | Structure of the Clade 2 C. difficile TcdB in complex with its receptor TFPI | | Descriptor: | Isoform Beta of Tissue factor pathway inhibitor, Toxin B | | Authors: | Luo, J, Yang, Q, Zhang, X, Zhang, Y, Wan, L, Li, Y, Tao, L. | | Deposit date: | 2021-08-05 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile.
Cell, 185, 2022
|
|
6AR6
 
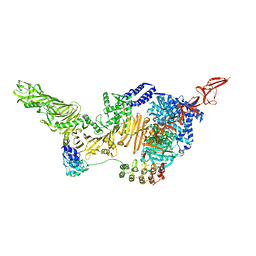 | |
6OQ5
 
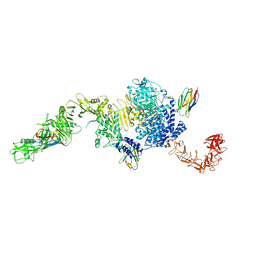 | | Structure of the full-length Clostridium difficile toxin B in complex with 3 VHHs | | Descriptor: | 5D, 7F, E3, ... | | Authors: | Chen, P, Lam, K, Jin, R. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structure of the full-length Clostridium difficile toxin B.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8QEO
 
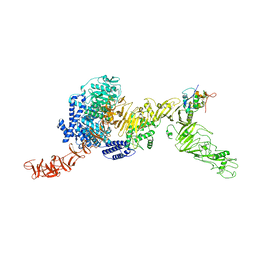 | |
8QEN
 
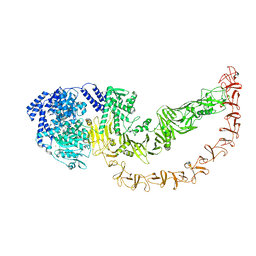 | |
8JHZ
 
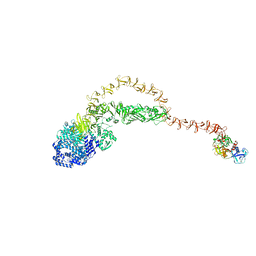 | | Cryo-EM structure of the TcsH-TMPRSS2 complex | | Descriptor: | Hemorrhagic toxin, Transmembrane protease serine 2, ZINC ION | | Authors: | Zhou, R, Liang, T, Zhan, X. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of TMPRSS2 recognition by Paeniclostridium sordellii hemorrhagic toxin.
Nat Commun, 15, 2024
|
|
8JB5
 
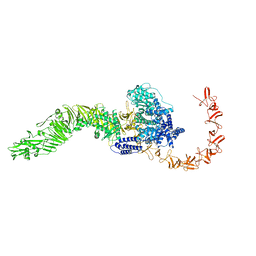 | |
8X2I
 
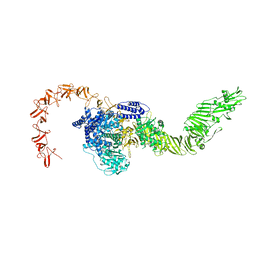 | |
8X2H
 
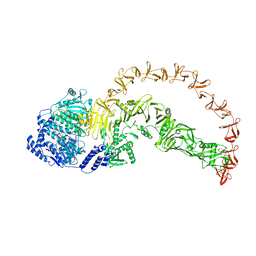 | |
8Y9C
 
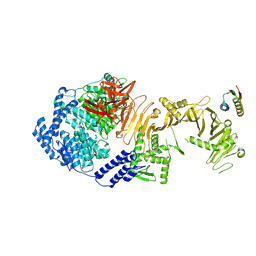 | | De novo design mini-binder in complex with TcdB4 | | Descriptor: | De novo design Minibinder, Toxin B, ZINC ION | | Authors: | Lv, X.C, Lu, P.L. | | Deposit date: | 2024-02-06 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | De novo design of mini-protein binders broadly neutralizing Clostridioides difficile toxin B variants.
Nat Commun, 15, 2024
|
|
8Y9B
 
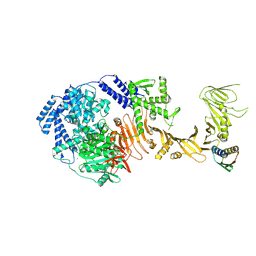 | | TcdB1 in complex with mini-binder | | Descriptor: | De novo design mini-binder, Toxin B, ZINC ION | | Authors: | Lv, X.C, Lu, P.L. | | Deposit date: | 2024-02-06 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | De novo design of mini-protein binders broadly neutralizing Clostridioides difficile toxin B variants.
Nat Commun, 15, 2024
|
|
7ML7
 
 | |
