1RL4
 
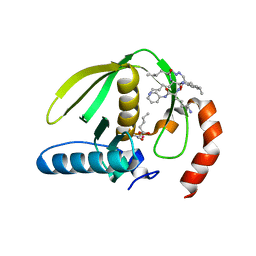 | | Plasmodium falciparum peptide deformylase complex with inhibitor | | Descriptor: | (2R)-2-{[FORMYL(HYDROXY)AMINO]METHYL}HEXANOIC ACID, 2-{N'-[2-(5-AMINO-1-PHENYLCARBAMOYL-PENTYLCARBAMOYL)-HEXYL]-HYDRAZINOMETHYL}-HEXANOIC ACID(5-AMINO-1-PHENYLCARBAMOYL-PENTYL)-AMIDE, COBALT (II) ION, ... | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G.J. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase.
Protein Sci., 13, 2004
|
|
1RL5
 
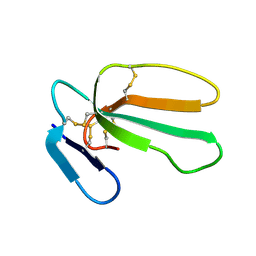 | | NMR structure with tightly bound water molecule of cytotoxin I from Naja oxiana in aqueous solution (major form) | | Descriptor: | Cytotoxin 1 | | Authors: | Dubinnyi, M.A, Pustovalova, Y.E, Dubovskii, P.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2003-11-25 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins.
Biochem.J., 387, 2005
|
|
1RL6
 
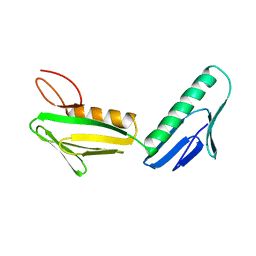 | | RIBOSOMAL PROTEIN L6 | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L6) | | Authors: | Golden, B.L, Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1999-01-14 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ribosomal protein L6: structural evidence of gene duplication from a primitive RNA binding protein.
EMBO J., 12, 1993
|
|
1RL8
 
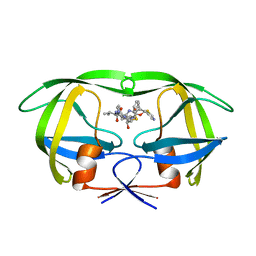 | | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir | | Descriptor: | RITONAVIR, protease RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Sedlacek, J, Konvalinka, J, Fabry, M, Horejsi, M. | | Deposit date: | 2003-11-25 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of resistant strain of hiv-1 protease(v82a mutant) with ritonavir
To be Published, 2005
|
|
1RL9
 
 | | Crystal structure of Creatine-ADP arginine kinase ternary complex | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, ... | | Authors: | Azzi, A, Clark, S.A, Ellington, R.W, Chapman, M.S. | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The role of phosphagen specificity loops in arginine kinase.
Protein Sci., 13, 2004
|
|
1RLA
 
 | | THREE-DIMENSIONAL STRUCTURE OF RAT LIVER ARGINASE, THE BINUCLEAR MANGANESE METALLOENZYME OF THE UREA CYCLE | | Descriptor: | ARGINASE, MANGANESE (II) ION | | Authors: | Kanyo, Z, Scolnick, L, Ash, D, Christianson, D.W. | | Deposit date: | 1996-08-15 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a unique binuclear manganese cluster in arginase.
Nature, 383, 1996
|
|
1RLB
 
 | |
1RLC
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
1RLD
 
 | | SOLID-STATE PHASE TRANSITION IN THE CRYSTAL STRUCTURE OF RIBULOSE 1,5-BIPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE | | Descriptor: | RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Eisenberg, D. | | Deposit date: | 1993-12-10 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solid-state phase transition in the crystal structure of ribulose 1,5-bisphosphate carboxylase/oxygenase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1RLF
 
 | | STRUCTURE DETERMINATION OF THE RAS-BINDING DOMAIN OF THE RAL-SPECIFIC GUANINE NUCLEOTIDE EXCHANGE FACTOR RLF, NMR, 10 STRUCTURES | | Descriptor: | RLF | | Authors: | Esser, D, Bauer, B, Wolthuis, R.M.F, Wittinghofer, A, Cool, R.H, Bay, P. | | Deposit date: | 1998-07-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the Ras-binding domain of the Ral-specific guanine nucleotide exchange factor Rlf.
Biochemistry, 37, 1998
|
|
1RLG
 
 | | Molecular basis of Box C/D RNA-protein interaction: co-crystal structure of the Archaeal sRNP intiation complex | | Descriptor: | 25-MER, 50S ribosomal protein L7Ae | | Authors: | Moore, T, Zhang, Y, Fenley, M.O, Li, H. | | Deposit date: | 2003-11-25 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis of Box C/D RNA-Protein Interactions; Cocrystal Structure of Archaeal L7Ae and a Box C/D RNA.
STRUCTURE, 12, 2004
|
|
1RLH
 
 | | Structure of a conserved protein from Thermoplasma acidophilum | | Descriptor: | SODIUM ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a conserved protein from T. acidophilum
To be Published
|
|
1RLI
 
 | |
1RLJ
 
 | | Structural Genomics, a Flavoprotein NrdI from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, IODIDE ION, NrdI protein | | Authors: | Wu, R, Zhang, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.5A crystal structure of a thioredoxin-like protein NrdI from Bacillus subtilis
To be Published
|
|
1RLK
 
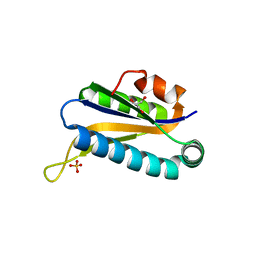 | | Structure of Conserved Protein of Unknown Function TA0108 from Thermoplasma acidophilum | | Descriptor: | GLYCEROL, Hypothetical protein Ta0108, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of conserved hypothetical protein TA0108 from Thermoplasma acidophilum
To be Published
|
|
1RLM
 
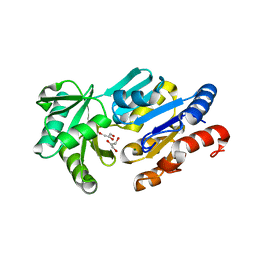 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLO
 
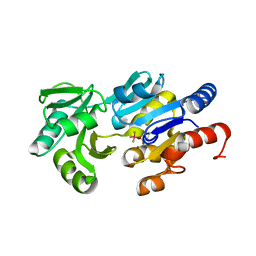 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLP
 
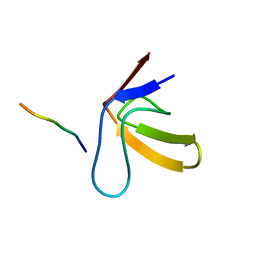 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RLQ
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RLR
 
 | |
1RLS
 
 | | CRYSTAL STRUCTURE OF RNASE T1 COMPLEXED WITH THE PRODUCT NUCLEOTIDE 3'-GMP. STRUCTURAL EVIDENCE FOR DIRECT INTERACTION OF HISTIDINE 40 AND GLUTAMIC ACID 58 WITH THE 2'-HYDROXYL GROUP OF RIBOSE | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Gohda, K, Oka, K.-I, Tomita, K.-I, Hakoshima, T. | | Deposit date: | 1994-03-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of RNase T1 complexed with the product nucleotide 3'-GMP. Structural evidence for direct interaction of histidine 40 and glutamic acid 58 with the 2'-hydroxyl group of the ribose.
J.Biol.Chem., 269, 1994
|
|
1RLT
 
 | | Transition State Analogue of ybiV from E. coli K12 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLU
 
 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein ftsZ, GLYCEROL | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-11-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
1RLV
 
 | |
1RLW
 
 | |
