1R71
 
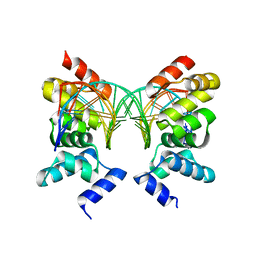 | | Crystal Structure of the DNA binding domain of KorB in complex with the operator DNA | | Descriptor: | 5'-D(*AP*(BRU)P*TP*TP*TP*AP*GP*CP*GP*GP*CP*TP*AP*AP*AP*AP*G)-3', 5'-D(*CP*(BRU)P*TP*TP*TP*AP*GP*CP*CP*GP*CP*TP*AP*AP*AP*AP*(BRU))-3', Transcriptional repressor protein korB | | Authors: | Khare, D, Ziegelin, G, Lanka, E, Heinemann, U. | | Deposit date: | 2003-10-17 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1R73
 
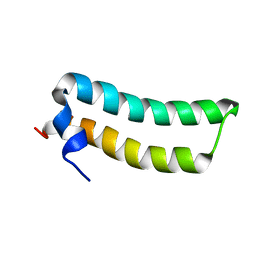 | | Solution Structure of TM1492, the L29 ribosomal protein from Thermotoga maritima | | Descriptor: | 50S ribosomal protein L29 | | Authors: | Peti, W, Etezady-Esfarjani, T, Herrmann, T, Klock, H.E, Lesley, S.A, Wuethrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-10-17 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR for structural proteomics of Thermotoga maritima: Screening and structure determination
J.STRUCT.FUNCT.GENOM., 5, 2004
|
|
1R74
 
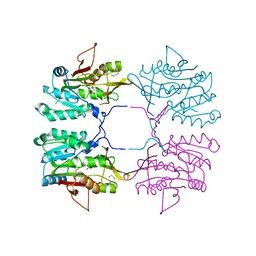 | | Crystal Structure of Human Glycine N-Methyltransferase | | Descriptor: | BETA-MERCAPTOETHANOL, CITRIC ACID, Glycine N-methyltransferase | | Authors: | Pakhomova, S, Luka, Z, Wagner, C, Newcomer, M.E. | | Deposit date: | 2003-10-17 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes.
Proteins, 57, 2004
|
|
1R75
 
 | |
1R76
 
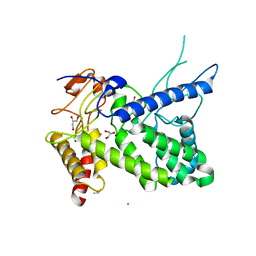 | | Structure of a pectate lyase from Azospirillum irakense | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Novoa de Armas, H, Verboven, C, De Ranter, C, Desair, J, Vande Broek, A, Vanderleyden, J, Rabijns, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Azospirillum irakense pectate lyase displays a toroidal fold.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R77
 
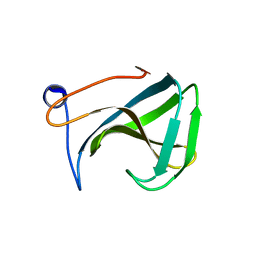 | | Crystal structure of the cell wall targeting domain of peptidylglycan hydrolase ALE-1 | | Descriptor: | Cell Wall Targeting Domain of Glycylglycine Endopeptidase ALE-1 | | Authors: | Lu, J.Z, Fujiwara, T, Komatsuzawa, H, Sugai, M, Sakon, J. | | Deposit date: | 2003-10-20 | | Release date: | 2005-04-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cell Wall-targeting Domain of Glycylglycine Endopeptidase Distinguishes among Peptidoglycan Cross-bridges.
J.Biol.Chem., 281, 2006
|
|
1R78
 
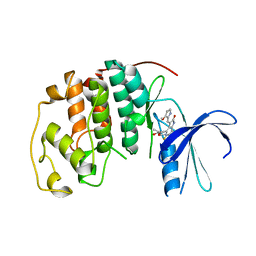 | | CDK2 complex with a 4-alkynyl oxindole inhibitor | | Descriptor: | 4-((3R,4S,5R)-4-AMINO-3,5-DIHYDROXY-HEX-1-YNYL)-5-FLUORO-3-[1-(3-METHOXY-1H-PYRROL-2-YL)-METH-(Z)-YLIDENE]-1,3-DIHYDRO-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Luk, K.-C, Simcox, M.E, Schutt, A, Rowan, K, Thompson, T, Chen, Y, Kammlott, U, DePinto, W, Dunten, P, Dermatakis, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new series of potent oxindole inhibitors of CDK2
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R79
 
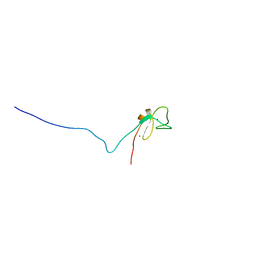 | | Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta | | Descriptor: | Diacylglycerol kinase, delta, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-21 | | Release date: | 2004-04-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The C1 Domain of The Human Diacylglycerol Kinase Delta
To be Published
|
|
1R7A
 
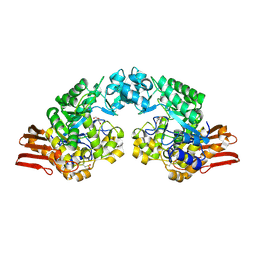 | | Sucrose Phosphorylase from Bifidobacterium adolescentis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, sucrose phosphorylase | | Authors: | Sprogoe, D, van den Broek, L.A.M, Mirza, O, Kastrup, J.S, Voragen, A.G.J, Gajhede, M, Skov, L.K. | | Deposit date: | 2003-10-21 | | Release date: | 2004-02-10 | | Last modified: | 2014-11-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis.
Biochemistry, 43, 2004
|
|
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7D
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 51 structures, sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7H
 
 | |
1R7I
 
 | | HMG-CoA Reductase from P. mevalonii, native structure at 2.2 angstroms resolution. | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, SULFATE ION | | Authors: | Watson, J.M, Steussy, C.N, Burgner, J.W, Lawrence, C.M, Tabernero, L, Rodwell, V.W, Stauffacher, C.V. | | Deposit date: | 2003-10-21 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Investigations of the Basis for Stereoselectivity from the Binary Complex of HMG-COA Reductase.
To be Published
|
|
1R7J
 
 | | Crystal structure of the DNA-binding protein Sso10a from Sulfolobus solfataricus | | Descriptor: | Conserved hypothetical protein Sso10a | | Authors: | Chen, L, Chen, L.R, Zhou, X.E, Wang, Y, Kahsai, M.A, Clark, A.T, Edmondson, S.P, Liu, Z.-J, Rose, J.P, Wang, B.C, Shriver, J.W, Meehan, E.J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The hyperthermophile protein Sso10a is a dimer of winged helix DNA-binding domains linked by an antiparallel coiled coil rod.
J.Mol.Biol., 341, 2004
|
|
1R7L
 
 | | 2.0 A Crystal Structure of a Phage Protein from Bacillus cereus ATCC 14579 | | Descriptor: | Phage protein | | Authors: | Zhang, R, Joachimiak, G, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of phage protein BC1872 from Bacillus cereus, a singleton with new fold
Proteins, 64, 2006
|
|
1R7M
 
 | | The homing endonuclease I-SceI bound to its DNA recognition region | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*AP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*TP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Moure, C.M, Gimble, F.S, Quiocho, F.A. | | Deposit date: | 2003-10-21 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity
J.Mol.Biol., 334, 2003
|
|
1R7O
 
 | |
1R7R
 
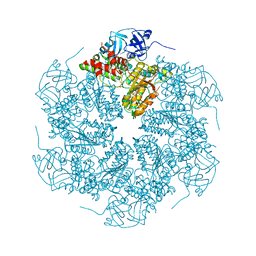 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
1R7S
 
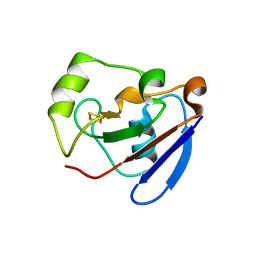 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1R7T
 
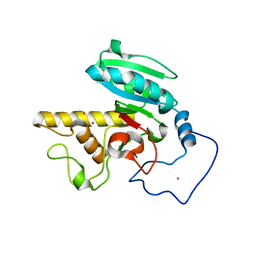 | | Glycosyltransferase A in complex with 3-deoxy-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7U
 
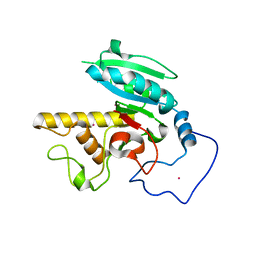 | | Glycosyltransferase B in complex with 3-deoxy-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-galactosyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
1R7V
 
 | | Glycosyltransferase A in complex with 3-amino-acceptor analog inhibitor | | Descriptor: | Glycoprotein-fucosylgalactoside alpha-N-acetylgalactosaminyltransferase, MERCURY (II) ION, alpha-L-fucopyranose-(1-2)-hexyl 3-amino-3-deoxy-beta-D-galactopyranoside | | Authors: | Nguyen, H.P, Seto, N.O.L, Cai, Y, Leinala, E.K, Borisova, S.N, Palcic, M.M, Evans, S.V. | | Deposit date: | 2003-10-22 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The influence of an intramolecular hydrogen bond in differential recognition of inhibitory acceptor analogs by human ABO(H) blood group A and B glycosyltransferases
J.Biol.Chem., 278, 2003
|
|
