1MA6
 
 | |
1MA7
 
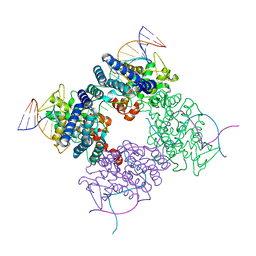 | |
1MA8
 
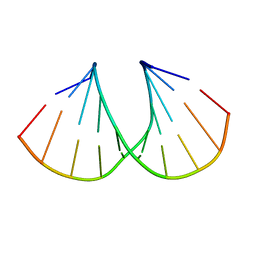 | | A-DNA decamer GCGTA(UMS)ACGC with incorporated 2'-methylseleno-uridine | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*UMSP*AP*CP*GP*C)-3' | | Authors: | Teplova, M, Wilds, C.J, Wawrzak, Z, Tereshko, V, Du, Q, Carrasco, N, Huang, Z, Egli, M. | | Deposit date: | 2002-08-01 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle
BIOCHIMIE, 84, 2002
|
|
1MA9
 
 | | Crystal structure of the complex of human vitamin D binding protein and rabbit muscle actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Alpha Skeletal Muscle, ... | | Authors: | Verboven, C, Bogaerts, I, Waelkens, E, Rabijns, A, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2002-08-02 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Actin-DBP: the perfect structural fit?
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1MAA
 
 | | MOUSE ACETYLCHOLINESTERASE CATALYTIC DOMAIN, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Taylor, P, Bougis, P.E, Marchot, P. | | Deposit date: | 1998-11-04 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of mouse acetylcholinesterase. A peripheral site-occluding loop in a tetrameric assembly.
J.Biol.Chem., 274, 1999
|
|
1MAB
 
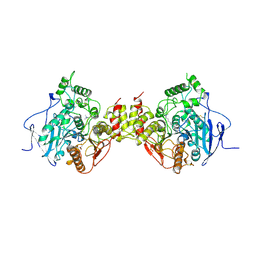
 | | RAT LIVER F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bianchet, M.A, Amzel, L.M. | | Deposit date: | 1998-08-06 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8-A structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1MAC
 
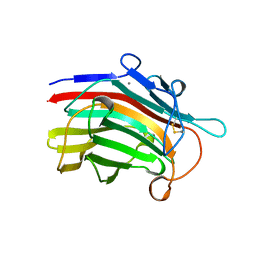 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
1MAE
 
 | |
1MAF
 
 | |
1MAG
 
 | |
1MAH
 
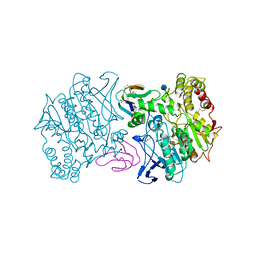 | | FASCICULIN2-MOUSE ACETYLCHOLINESTERASE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, FASCICULIN 2 | | Authors: | Bourne, Y, Taylor, P, Marchot, P. | | Deposit date: | 1995-11-21 | | Release date: | 1996-04-03 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Acetylcholinesterase inhibition by fasciculin: crystal structure of the complex.
Cell(Cambridge,Mass.), 83, 1995
|
|
1MAI
 
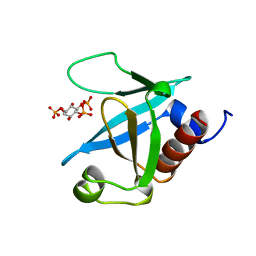 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM PHOSPHOLIPASE C DELTA IN COMPLEX WITH INOSITOL TRISPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOLIPASE C DELTA-1 | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1996-05-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain.
Cell(Cambridge,Mass.), 83, 1995
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAL
 
 | |
1MAM
 
 | |
1MAP
 
 | |
1MAQ
 
 | |
1MAR
 
 | |
1MAS
 
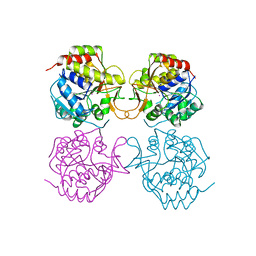 | | PURINE NUCLEOSIDE HYDROLASE | | Descriptor: | INOSINE-URIDINE NUCLEOSIDE N-RIBOHYDROLASE, POTASSIUM ION | | Authors: | Degano, M, Gopaul, D.N, Scapin, G, Schramm, V.L, Sacchettini, J.C. | | Deposit date: | 1995-12-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of the inosine-uridine nucleoside N-ribohydrolase from Crithidia fasciculata.
Biochemistry, 35, 1996
|
|
1MAT
 
 | |
1MAU
 
 | | Crystal structure of Tryptophanyl-tRNA Synthetase Complexed with ATP and Tryptophanamide in a Pre-Transition state Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, GLYCEROL, ... | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1MAV
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 6.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1MAW
 
 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with ATP in an Open Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, TRYPTOPHAN-TRNA LIGASE | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1MAX
 
 | | BETA-TRYPSIN PHOSPHONATE INHIBITED | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, [N-(BENZYLOXYCARBONYL)AMINO](4-AMIDINOPHENYL)METHANE-PHOSPHONATE | | Authors: | Bertrand, J, Oleksyszyn, J, Kam, C, Boduszek, B, Presnell, S, Plaskon, R, Suddath, F, Powers, J, Williams, L. | | Deposit date: | 1996-02-06 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of trypsin and thrombin by amino(4-amidinophenyl)methanephosphonate diphenyl ester derivatives: X-ray structures and molecular models.
Biochemistry, 35, 1996
|
|
