1K70
 
 | | The Structure of Escherichia coli Cytosine Deaminase bound to 4-Hydroxy-3,4-Dihydro-1H-Pyrimidin-2-one | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1K72
 
 | | The X-ray Crystal Structure Of Cel9G Complexed With cellotriose | | Descriptor: | CALCIUM ION, Endoglucanase 9G, GLYCEROL, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1K73
 
 | | Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit | | Descriptor: | 23S RRNA, 5S RRNA, ANISOMYCIN, ... | | Authors: | Hansen, J, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
1K74
 
 | | The 2.3 Angstrom resolution crystal structure of the heterodimer of the human PPARgamma and RXRalpha ligand binding domains respectively bound with GW409544 and 9-cis retinoic acid and co-activator peptides. | | Descriptor: | (9cis)-retinoic acid, 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor gamma, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K75
 
 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K76
 
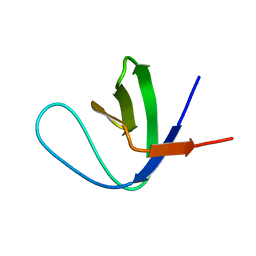 | | Solution Structure of the C-terminal Sem-5 SH3 Domain (Minimized Average Structure) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J, Volk, D, Luxon, B, Gorenstein, D, Hilser, V. | | Deposit date: | 2001-10-18 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1K77
 
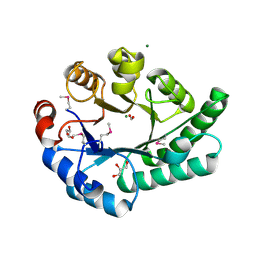 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1K78
 
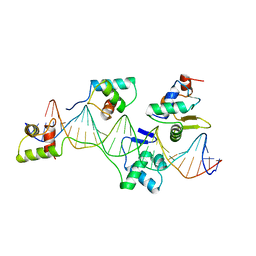 | | Pax5(1-149)+Ets-1(331-440)+DNA | | Descriptor: | C-ets-1 Protein, Paired Box Protein Pax5, Pax5/Ets Binding Site on the mb-1 promoter | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K79
 
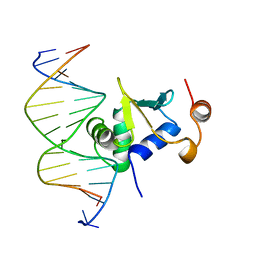 | | Ets-1(331-440)+GGAA duplex | | Descriptor: | C-ets-1 protein, DNA (5'-D(*CP*AP*CP*AP*TP*TP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*AP*AP*TP*GP*T)-3') | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K7A
 
 | | Ets-1(331-440)+GGAG duplex | | Descriptor: | C-ets-1 Protein, DNA (5'-D(*CP*AP*CP*AP*TP*CP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*GP*AP*TP*GP*T)-3') | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K7B
 
 | | NMR Solution Structure of sTva47, the Viral-Binding Domain of Tva | | Descriptor: | SUBGROUP A ROUS SARCOMA VIRUS RECEPTOR PG800 AND PG950 | | Authors: | Tonelli, M, Peters, R.J, James, T.L, Agard, D.A. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-19 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus.
FEBS Lett., 509, 2001
|
|
1K7C
 
 | |
1K7D
 
 | | Penicillin Acylase with Phenyl Proprionic Acid | | Descriptor: | CALCIUM ION, Penicillin Acylase alpha subunit, R-2-PHENYL-PROPRIONIC ACID, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-19 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1K7E
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K7F
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]VALINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]VALINE ACID, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K7G
 
 | | PrtC from Erwinia chrysanthemi | | Descriptor: | CALCIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Hege, T, Baumann, U. | | Deposit date: | 2001-10-19 | | Release date: | 2002-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1K7H
 
 | | CRYSTAL STRUCTURE OF SHRIMP ALKALINE PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALKALINE PHOSPHATASE, MALEIC ACID, ... | | Authors: | De Backer, M.E, Mc Sweeney, S, Rasmussen, H.B, Riise, B.W, Lindley, P, Hough, E. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.9 A Crystal Structure of Heat-Labile Shrimp Alkaline Phosphatase
J.Mol.Biol., 318, 2002
|
|
1K7I
 
 | | PrtC from Erwinia chrysanthemi: Y228F mutant | | Descriptor: | CALCIUM ION, ZINC ION, secreted protease C | | Authors: | Baumann, U, Hege, T. | | Deposit date: | 2001-10-19 | | Release date: | 2002-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1K7J
 
 | | Structural Genomics, protein TF1 | | Descriptor: | Protein yciO, SULFATE ION | | Authors: | Zhang, R, Dementieva, I, Thorn, J, Donnelly, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Genomics, protein TF1
To be Published
|
|
1K7K
 
 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
1K7L
 
 | | The 2.5 Angstrom resolution crystal structure of the human PPARalpha ligand binding domain bound with GW409544 and a co-activator peptide. | | Descriptor: | 2-(1-METHYL-3-OXO-3-PHENYL-PROPYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator activated receptor alpha, YTTRIUM (III) ION, ... | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Moore, L.B, Collins, J.L, Oplinger, J.A, Kliewer, S.A, Gampe Jr, R.T, McKee, D.D, Moore, J.T, Willson, T.M. | | Deposit date: | 2001-10-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K7Q
 
 | | PrtC from Erwinia chrysanthemi: E189A mutant | | Descriptor: | CALCIUM ION, SECRETED PROTEASE C, ZINC ION | | Authors: | Baumann, U, Hege, T. | | Deposit date: | 2001-10-20 | | Release date: | 2002-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1K7S
 
 | | FhuD complexed with albomycin-delta 2 | | Descriptor: | DELTA-2-ALBOMYCIN A1, Ferrichrome-binding periplasmic protein | | Authors: | Clarke, T.E, Braun, V, Winkelmann, G, Tari, L.W, Vogel, H.J. | | Deposit date: | 2001-10-21 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin.
J.Biol.Chem., 277, 2002
|
|
1K7T
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,6Gal complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1K7U
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,4GlcNAc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
