1JBL
 
 | | Solution structure of SFTI-1, A cyclic trypsin inhibitor from sunflower seeds | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Korsinczky, M.L.J, Schirra, H.J, Rosengren, K.J, West, J, Condie, B.A, Otvos, L, Anderson, M.A, Craik, D.J. | | Deposit date: | 2001-06-05 | | Release date: | 2001-08-22 | | Last modified: | 2015-04-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant.
J.Mol.Biol., 311, 2001
|
|
1JBM
 
 | |
1JBN
 
 | | Solution structure of an acyclic permutant of SFTI-1, A trypsin inhibitor from sunflower seeds | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Korsinczky, M.L.J, Schirra, H.J, Rosengren, K.J, West, J, Condie, B.A, Otvos, L, Anderson, M.A, Craik, D.J. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-22 | | Last modified: | 2016-12-28 | | Method: | SOLUTION NMR | | Cite: | Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant.
J.Mol.Biol., 311, 2001
|
|
1JBO
 
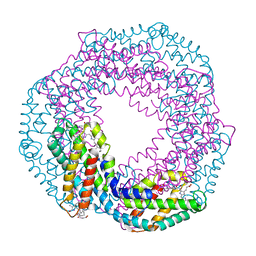 | | The 1.45A Three-Dimensional Structure of c-Phycocyanin from the Thermophylic Cyanobacterium Synechococcus elongatus | | Descriptor: | C-Phycocyanin alpha chain, C-Phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Nield, J, Rizkallah, P.J, Barber, J, Chayen, N.E. | | Deposit date: | 2002-05-02 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The 1.45A three-dimensional structure of C-phycocyanin from the
thermophilic cyanobacterium Synechococcus elongatus
J.STRUCT.BIOL., 141, 2003
|
|
1JBP
 
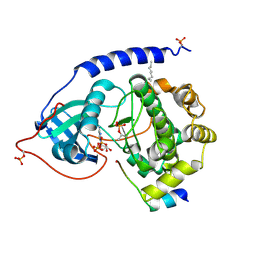 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Substrate Peptide, ADP and Detergent | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, MUSCLE/BRAIN FORM, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
1JBQ
 
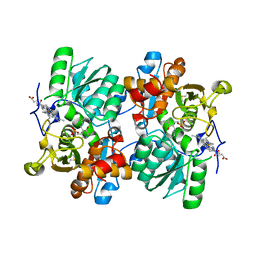 | | STRUCTURE OF HUMAN CYSTATHIONINE BETA-SYNTHASE: A UNIQUE PYRIDOXAL 5'-PHOSPHATE DEPENDENT HEMEPROTEIN | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meier, M, Janosik, M, Kery, V, Kraus, J.P, Burkhard, P. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein.
EMBO J., 20, 2001
|
|
1JBR
 
 | | Crystal Structure of the Ribotoxin Restrictocin and a 31-mer SRD RNA Inhibitor | | Descriptor: | 31-mer SRD RNA analog, 5'-R(*GP*CP*GP*CP*UP*CP*CP*UP*CP*AP*GP*UP*AP*CP*GP*AP*GP*(A23))-3', 5'-R(*GP*GP*AP*AP*CP*CP*GP*GP*AP*GP*CP*GP*C)-3', ... | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBS
 
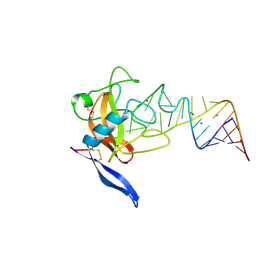 | | Crystal structure of ribotoxin restrictocin and a 29-mer SRD RNA analog | | Descriptor: | 29-mer sarcin/ricin domain RNA analog, POTASSIUM ION, restrictocin | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBT
 
 | | CRYSTAL STRUCTURE OF RIBOTOXIN RESTRICTOCIN COMPLEXED WITH A 29-MER SARCIN/RICIN DOMAIN RNA ANALOG | | Descriptor: | 29-MER SARCIN/RICIN DOMAIN RNA ANALOG, POTASSIUM ION, RESTRICTOCIN | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBU
 
 | | Coagulation Factor VII Zymogen (EGF2/Protease) in Complex with Inhibitory Exosite Peptide A-183 | | Descriptor: | BENZAMIDINE, COAGULATION FACTOR VII, Peptide exosite inhibitor A-183, ... | | Authors: | Eigenbrot, C, Kirchhofer, D, Dennis, M.S, Santell, L, Lazarus, R.A, Stamos, J, Ultsch, M.H. | | Deposit date: | 2001-06-06 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The factor VII zymogen structure reveals reregistration of beta strands during activation.
Structure, 9, 2001
|
|
1JBV
 
 | | FPGS-AMPPCP complex | | Descriptor: | FOLYLPOLYGLUTAMATE SYNTHASE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
1JBW
 
 | | FPGS-AMPPCP-folate complex | | Descriptor: | 5,10-METHYLENE-6-HYDROFOLIC ACID, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, FOLYLPOLYGLUTAMATE SYNTHASE, ... | | Authors: | Sun, X, Cross, J.A, Bognar, A.L, Baker, E.N, Smith, C.A. | | Deposit date: | 2001-06-06 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Folate-binding triggers the activation of folylpolyglutamate synthetase.
J.Mol.Biol., 310, 2001
|
|
1JBY
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT LOW PH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1JBZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A DUAL-WAVELENGTH EMISSION GREEN FLUORESCENT PROTEIN VARIANT AT HIGH PH | | Descriptor: | 1,2-ETHANEDIOL, GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | Hanson, G.T, McAnaney, T.B, Park, E.S, Rendell, M.E.P, Yarbrough, D.K, Chu, S, Xi, L, Boxer, S.G, Montrose, M.H, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Variants as Ratiometric Dual Emission pH Sensors. 1. Structural Characterization and Preliminary Application.
Biochemistry, 41, 2002
|
|
1JC0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A REDUCED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
1JC1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A REDOX-SENSITIVE GREEN FLUORESCENT PROTEIN VARIANT IN A OXIDIZED FORM | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Hanson, G.T, Aggeler, R, Oglesbee, D, Cannon, M, Capaldi, R.A, Tsien, R.Y, Remington, S.J. | | Deposit date: | 2001-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators.
J.Biol.Chem., 279, 2004
|
|
1JC2
 
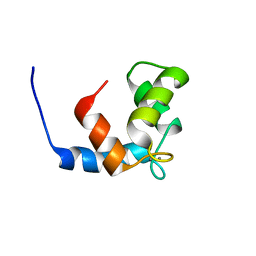 | | COMPLEX OF THE C-DOMAIN OF TROPONIN C WITH RESIDUES 1-40 OF TROPONIN I | | Descriptor: | CALCIUM ION, TROPONIN C, SKELETAL MUSCLE | | Authors: | Mercier, P, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 2001-06-07 | | Release date: | 2001-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and thermodynamics of the structural domain of troponin C in complex with the regulatory peptide 1-40 of troponin I
Biochemistry, 40, 2001
|
|
1JC4
 
 | | Crystal Structure of Se-Met Methylmalonyl-CoA Epimerase | | Descriptor: | Methylmalonyl-CoA epimerase, SULFATE ION | | Authors: | Mc Carthy, A.A, Baker, H.M, Shewry, S.C, Patchett, M.L, Baker, E.N. | | Deposit date: | 2001-06-07 | | Release date: | 2001-07-11 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
Structure, 9, 2001
|
|
1JC5
 
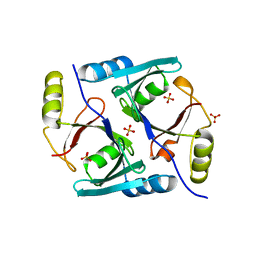 | | Crystal Structure of Native Methylmalonyl-CoA Epimerase | | Descriptor: | Methylmalonyl-CoA Epimerase, SULFATE ION | | Authors: | Mc Carthy, A.A, Baker, H.M, Shewry, S.C, Patchett, M.L, Baker, E.N. | | Deposit date: | 2001-06-07 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold.
Structure, 9, 2001
|
|
1JC6
 
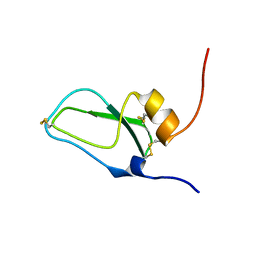 | | SOLUTION STRUCTURE OF BUNGARUS FACIATUS IX, A KUNITZ-TYPE CHYMOTRYPSIN INHIBITOR | | Descriptor: | VENOM BASIC PROTEASE INHIBITORS IX AND VIIIB | | Authors: | Chen, C, Hsu, C.H, Su, N.Y, Chiou, S.H, Wu, S.H. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Kunitz-type chymotrypsin inhibitor isolated from the elapid snake Bungarus fasciatus
J.BIOL.CHEM., 276, 2001
|
|
1JC7
 
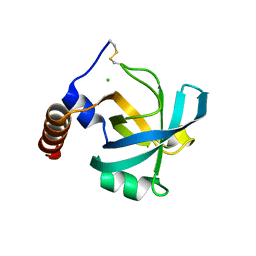 | |
1JC8
 
 | | Solution structure of lactam analogue (DDap) of gp41 600-612 loop of HIV | | Descriptor: | DDap: (ACE)IWGDSGKLI(DNP)TTA ANALOGUE OF HIV GP41 | | Authors: | Phan Chan Du, A, Limal, D, Semetey, V, Dali, H, Jolivet, M, Desgranges, C, Cung, M.T, Briand, J.P, Petit, M.C, Muller, S. | | Deposit date: | 2001-06-08 | | Release date: | 2003-07-01 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Structural and immunological characterisation of heteroclitic peptide analogues corresponding to the 600-612 region of the HIV envelope gp41 glycoprotein.
J.Mol.Biol., 323, 2002
|
|
1JC9
 
 | | TACHYLECTIN 5A FROM TACHYPLEUS TRIDENTATUS (JAPANESE HORSESHOE CRAB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, techylectin-5A | | Authors: | Kairies, N, Beisel, H.-G, Fuentes-Prior, P, Tsuda, R, Muta, T, Iwanaga, S, Bode, W, Huber, R, Kawabata, S. | | Deposit date: | 2001-06-08 | | Release date: | 2001-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1JCC
 
 | | Crystal Structure of a Novel Alanine-Zipper Trimer at 1.7 A Resolution, V13A,L16A,V20A,L23A,V27A,M30A,V34A mutations | | Descriptor: | MAJOR OUTER MEMBRANE LIPOPROTEIN, ZINC ION | | Authors: | Liu, J, Dai, J, Lu, M. | | Deposit date: | 2001-06-08 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Zinc-Mediated Helix Capping in A Triple-Helical Protein
Biochemistry, 42, 2003
|
|
