9ICD
 
 | | CATALYTIC MECHANISM OF NADP+-DEPENDENT ISOCITRATE DEHYDROGENASE: IMPLICATIONS FROM THE STRUCTURES OF MAGNESIUM-ISOCITRATE AND NADP+ COMPLEXES | | Descriptor: | ISOCITRATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hurley, J.H, Dean, A.M, Koshland Jr, D.E, Stroud, R.M. | | Deposit date: | 1991-07-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes.
Biochemistry, 30, 1991
|
|
6XXY
 
 | | Crystal structure of Haemophilus influenzae 3-isopropylmalate dehydrogenase in complex with O-isobutenyl oxalylhydroxamate. | | Descriptor: | 2-(2-methylprop-2-enoxyamino)-2-oxidanylidene-ethanoic acid, 3-isopropylmalate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Miggiano, R, Rossi, F, Martignon, S, Rizzi, M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Haemophilus influenzae 3-isopropylmalate dehydrogenase (LeuB) in complex with the inhibitor O-isobutenyl oxalylhydroxamate.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
4F7I
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus in complex with IPM, Mn and NADH | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Pallo, A, Graczer, E, Zavodszky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic basis of isopropylmalate dehydrogenase enzyme catalysis.
Febs J., 281, 2014
|
|
2G4O
 
 | | anomalous substructure of 3-ISOPROPYLMALATE DEHYDROGENASE | | Descriptor: | 3-isopropylmalate dehydrogenase, CHLORIDE ION, SULFATE ION | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
4WUO
 
 | | Structure of the E270A Mutant Isopropylmalate dehydrogenase from Thermus thermophilus in complex with IPM, Mn and NADH | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, ETHANOL, ... | | Authors: | Pallo, A, Graczer, E, Olah, J, Szimler, T, Konarev, P.V, Svergun, D.I, Merli, A, Zavodszky, P, Vas, M, Weiss, M.S. | | Deposit date: | 2014-11-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glutamate 270 plays an essential role in K(+)-activation and domain closure of Thermus thermophilus isopropylmalate dehydrogenase.
Febs Lett., 589, 2015
|
|
5TQH
 
 | | IDH1 R132H mutant in complex with IDH889 | | Descriptor: | (4S)-3-[2-({(1S)-1-[5-(4-fluoro-3-methylphenyl)pyrimidin-2-yl]ethyl}amino)pyrimidin-4-yl]-4-(propan-2-yl)-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 3-Pyrimidin-4-yl-oxazolidin-2-ones as Allosteric and Mutant Specific Inhibitors of IDH1.
ACS Med Chem Lett, 8, 2017
|
|
2IV0
 
 | | Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure analysis and engineering of chimers | | Descriptor: | CHLORIDE ION, ISOCITRATE DEHYDROGENASE, ZINC ION | | Authors: | Stokke, R, Karlstrom, M, Yang, N, Leiros, I, Ladenstein, R, Birkeland, N.K, Steen, I.H. | | Deposit date: | 2006-06-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thermal Stability of Isocitrate Dehydrogenase from Archaeoglobus Fulgidus Studied by Crystal Structure Analysis and Engineering of Chimers
Extremophiles, 11, 2007
|
|
5DE1
 
 | | Crystal structure of human IDH1 in complex with GSK321A | | Descriptor: | (7R)-1-(4-fluorobenzyl)-N-{3-[(1S)-1-hydroxyethyl]phenyl}-7-methyl-5-(1H-pyrrol-2-ylcarbonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridine-3-carboxamide, Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Concha, N.O, Smallwood, A, Qi, H. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New IDH1 mutant inhibitors for treatment of acute myeloid leukemia.
Nat.Chem.Biol., 11, 2015
|
|
8BAY
 
 | | Crystal Structure of IDH1 variant R132C S280F in complex with NADPH, Ca2+ and 3-butyl-2-oxoglutarate | | Descriptor: | (R)-3-butyl-2-oxopentanedioic acid, (S)-3-butyl-2-oxopentanedioic acid, CALCIUM ION, ... | | Authors: | Rabe, P, Schofield, C.J, Reinbold, R, Brewitz, L. | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Natural and synthetic 2-oxoglutarate derivatives are substrates for oncogenic variants of human isocitrate dehydrogenase 1 and 2.
J.Biol.Chem., 299, 2023
|
|
7ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
6U4J
 
 | | Crystal structure of IDH1 R132H mutant in complex with FT-2102 | | Descriptor: | 5-{[(1S)-1-(6-chloro-2-oxo-1,2-dihydroquinolin-3-yl)ethyl]amino}-1-methyl-6-oxo-1,6-dihydropyridine-2-carbonitrile, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Toms, A.V, Lin, J. | | Deposit date: | 2019-08-25 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Based Design and Identification of FT-2102 (Olutasidenib), a Potent Mutant-Selective IDH1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
5M2E
 
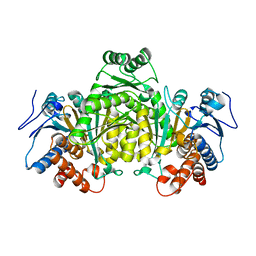 | |
4BNP
 
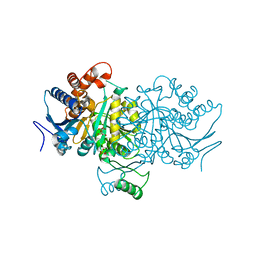 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with isocitrate and magnesium(II) | | Descriptor: | ISOCITRATE DEHYDROGENASE [NADP], ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Miller, S.P, Goncalves, S, Matias, P.M, Dean, A.M. | | Deposit date: | 2013-05-16 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a Transition State: Role of Lys100 in the Active Site of Isocitrate Dehydrogenase.
Chembiochem, 15, 2014
|
|
2QFY
 
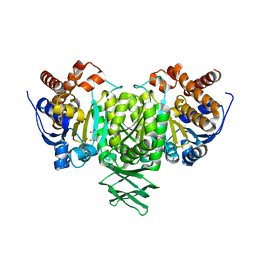 | |
3BLW
 
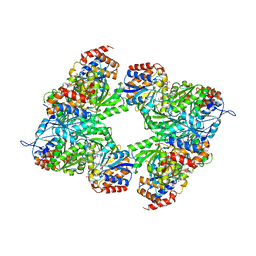 | | Yeast Isocitrate Dehydrogenase with Citrate and AMP Bound in the Regulatory Subunits | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, ... | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
4XRX
 
 | | Crystal structure of a metabolic reductase with (E)-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one | | Descriptor: | 5-[(E)-(1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl]pyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, B, Wu, F, Jiang, H, Kogiso, M, Yao, Y, Zhou, C, Li, X, Song, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds.
J.Med.Chem., 58, 2015
|
|
7PJN
 
 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and inhibitor DS-1001B | | Descriptor: | (E)-3-(1-(5-(2-fluoropropan-2-yl)-3-(2,4,6-trichlorophenyl)isoxazole-4-carbonyl)-3-methyl-1H-indol-4-yl)acrylic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J, Clifton, I.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
7PJM
 
 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
1W0D
 
 | | The high resolution structure of Mycobacterium tuberculosis LeuB (Rv2995c) | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Singh, R.K, Kefala, G, Janowski, R, Mueller-Dieckmann, C, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-06-03 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The High Resolution Structure of Leub (Rv2995C) from Mycobacterium Tuberculosis
J.Mol.Biol., 346, 2005
|
|
3BLV
 
 | | Yeast Isocitrate Dehydrogenase with Citrate Bound in the Regulatory Subunits | | Descriptor: | CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
5J32
 
 | | Isopropylmalate dehydrogenase in complex with isopropylmalate | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase 2, chloroplastic, ... | | Authors: | Jez, J.M, Lee, S.G. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
5J33
 
 | | Isopropylmalate dehydrogenase in complex with NAD+ | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, MAGNESIUM ION, ... | | Authors: | Jez, J.M, Lee, S.G. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | Structure and Mechanism of Isopropylmalate Dehydrogenase from Arabidopsis thaliana: INSIGHTS ON LEUCINE AND ALIPHATIC GLUCOSINOLATE BIOSYNTHESIS.
J.Biol.Chem., 291, 2016
|
|
4YB4
 
 | | Crystal structure of homoisocitrate dehydrogenase from Thermus thermophilus in complex with homoisocitrate, magnesium ion (II) and NADH | | Descriptor: | (1R,2S)-1-hydroxybutane-1,2,4-tricarboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Takahashi, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of homoisocitrate dehydrogenase from Thermus thermophilus in complex with homoisocitrate, magnesium(II) and NADH
To Be Published
|
|
1X0L
 
 | | Crystal structure of tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus | | Descriptor: | Homoisocitrate dehydrogenase | | Authors: | Miyazaki, J, Asada, K, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2005-03-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Tetrameric Homoisocitrate Dehydrogenase from an Extreme Thermophile, Thermus thermophilus: Involvement of Hydrophobic Dimer-Dimer Interaction in Extremely High Thermotolerance
J.Bacteriol., 187, 2005
|
|
